ÓÄÊ 636:636.018:575.174.4
CONCEPT AND PRINCIPLES OF GENETIC MONITORING FOR THE PURPOSE OF PRESERVATION in situ OF DOMESTICAL ANIMALS KINDS
Yu.A. Stolpovskii
The author discussed the concept of genetic monitoring of gene pool of domestical kinds which based on seven interconnected blocks which accumulates the fenetic, biochemical, cytogenetic, molecular, zootechnical, veterinary, mathematical and computer methods. The proposed scheme of the monitoring of agricultural populations is general multilevel system and satisfies the three basic requirements. In the first, its usage promotes to preservation of a qualitative variety and structure of genofonds of breeds (populations). Secondly, it permits during of genotype selection to use the data of genetic systems variability so, that to achieve their representativeness by the minimum quantity of animals. Thirdly, the monitoring methods have to supplement each other, combine relative simplicity and reproducibility with maximum effectiveness.
Key words: genetic monitoring, gene pool, polymorphism, DNA markers, rare and endangered breeds, preservation of breeds in situ.
Genetic monitoring of inter- and intrapopulation gene flow is the part of a bioecological monitoring, whose main targets are a population, species and ecosystem. When preservation in situ of breeds the domesticated species, genetic monitoring has specific features – the object of a long-term control are intra- and interbreed genetic diversity, evaluation and prognostication of its dynamics, determination of its optimum and limits of acceptable changes. The genetic polymorphism (of phenes, structural genes, multi-loci DNA sequences, chromosomal and genomic mutations) labels genetic structure of a breed, working as a "genetic reference point" for preservation of gene pool of rare and endangered breeds.
Currently, it’s common to consider genetic monitoring to be especially important in small groups of animals or in populations where one breed is being replaced by another one; though, the principles of such studies are not sufficiently developed. The author suggest that determination of main regulations, elements, structure, methods of genetic monitoring should be the key element in programs for preservation and improvement of the gene pool of domesticated animals.
When preservation of species in situ, the primary target is prevention of losses of specific gene complexes (the balanced system of genes) responsible for breed phenotypic characteristics associated with animals’ exterior peculiarities, productivity, vitality and resistance. These features are namely the ones distinguishing local breeds from the wide-spread foreign species, and they must be preserved in pedigree farms.
It is obvious that an accurate monitoring, systematic observation, development of forecasting methods, clear criteria and effective evaluation of the gene pool state are necessary for simulation of breeding process and selecting an optimal program for in situ preservation of endangered rare and small in number animal breeds. Genetic monitoring may be local (a herd or population of a certain breed) or global (the pedigree diversity of a region or country).
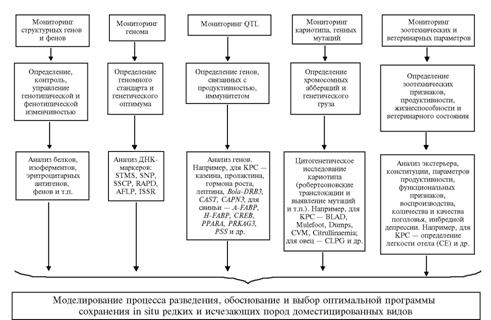
According to the authors’ concept, genetic monitoring of rare and endangered breeds and the breeds diversity is based on the seven units closely linked with each other and forming the integrated multi-level control system for observation, prognostication and management in preservation of the gene pool (see scheme).
|
The scheme of genetic monitoring in populations of rare and endangered breeds of animals |
Denotations to the scheme – rows from top to a bottom, left to right:
row 1, element 1: Monitoring of structural genes and phenes
row 1, element 2: Monitoring of the genome
row 1, element 3: Monitoring of QTLs
row 1, element 4: Monitoring of the caryotype and gene mutations
row 1, element 5: Monitoring of zootechnic and veterinary parameters
row 2, element 1: Determination, control and management of the genotypic and phenotypic diversity
row 2, element 2: Determination of the genomic standard and genetic optimum
row 2, element 3: Determination of the genes associated with productivity and immunity
row 2, element 4: Determination of chromosomal aberrations and the genetic load
row 2, element 5: Determination of zootechnic parameters, productivity, vitality and a veterinarian condition
row 3, element 1: Analysis of proteins, isoenzymes, erythrocyte antigens, phenes etc.
row 3, element 2: Analysis of DNA-markers, STMS, SNP, SSCP, RAPD, AFLP, ISSR
row 3, element 3: Analysis of genes. For example, in cattle – the genes for casein, prolactin, growth hormone, leptin, Bola-DRB3, CAST, CAN3, in swine – A-FABP, H-FABP, CREB, PPARA, PRKAG3, PSS, etc.
row 3, element 4: Cytogenetic investigation of the caryotype (Robertsonian translocations, identification of mutations, etc.). For example, in cattle – BLAD, Mulefoot, Dumps, CVM, Citrullinaemia, for sheep –CLPG, etc.
row 3, element 5: Analysis of exterior, physical build-up, productivity indices, functional parameters, reproduction, quantity and quality of progenies, the inbred depression. For example, in cattle – determination of calving ease (CE), etc.
the bottom element: Simulation of breeding processes, justification and selection of an optimal program for preservation in situ of rare and endangered domesticated species
The first unit - monitoring of phenes and structural genes. Phenetic method involves identification of genetically determined discrete (alternative) parameters (phenes) and their control (1, 2). In farm animals, there are dozens of discrete variations of such traits. The control of breed’s phenes is the way to maintain the uniqueness of its gene pool including a wide range of morphological variability, which is equal to preservation of genetic diversity in populations.
Domestic species have fairly high polymorphism of structural genes carrying information about amino acid sequence in a protein molecule. Evaluation of polymorphism in electrophoretic variants of proteins, enzymes and erythrocyte antigens helps to determine their distribution and to establish their involvement into evolution of species, to perform individual certification, to display genetically labeled lines and to control animals’ pedigree. The efficiency of breeding work can be increased by combined use of immunogenetic evaluation of individuals’ genetic resemblance and the analysis of their genotypes (3). Moreover, studying the protein polymorphism in different species, it has been shown its capabilities for determination of spatial and temporal allocations in genetic structure of populations, as well as for differentiation of commercial and local breeds (4, 5). During the propagation and transformation of monogenic loci responsible for immune and biochemical functions, their genotyping allows to identify the types of interbreed interactions.
The second unit - monitoring of the gene pool using DNA markers (monoloci - STMS, SNP, SSCP - and multiloci - RAPD, AFLP, ISSR). Microsatellites STMS (sequence tagged microsatellite site) are dispersed tandem repeat sequences (di-, tri-and tetra- nucleotides). A high level of polymorphism of numerous microsatellite loci revealed in genomes of domestic species was appraised by ISAG (International Society for Animal Genetics) who recommended using this method for individual genotyping and certification of breeds of cattle, horses and other species (6).
Identification of multiloci markers is based on polymerase chain reaction (PCR) using a single short primer having an arbitrary sequence (RAPD – the method of randomly amplified polymorphic DNA and its analogues), the primers with artificially added sequences (AFLP - amplified fragment length polymorphism) or the primers complementary to repetitive elements of genome, such as microsatellites (ISSR - inter simple sequence repeats). Among these methods, ISSR is the most attractive. The design of ISSR-markers doesn’t require prior knowledge of the nucleotide sequence in investigated DNA, which is important for little-studied species. The obtained patterns of PCR products are usually species- and breed-specific (the latter fact is very important). In comparative studies of multiloci spectra of DNA fragments flanked by segments of microsatellite loci (ISSR-PCR), it has been revealed the groups of markers, which can be used to describe a "genomic standard," "genomic profile", "breed genomic pattern" and "genetic optimum" of animal breeds preserved in situ (7).
The third unit – monitoring of key genes of quantitative traits (QTL - quantitative trait loci). It includes a search and typing of known and/or associated with productivity genes. For example, these are the genes for k- and b- casein (CSN1, CSN2), prolactin (PRL), a-lactalbumin (LALBA), growth hormone (GH), calpains – the cysteine proteases determining softness of meat in cattle (CAST), the genes affecting regeneration of muscles in sheep (CAPN3), fat characteristics in pigs (FABP), regulation factor of transcription involved in adipogenesis (CREB), a receptor that controls intracellular lipid metabolism (PPARA), the gene associated with glycogen content in muscles of pigs (PKAG3), the gene for stress syndrome (PSS) of pigs (RYR-1), etc. There can be also mentioned the genes controlling immune responses (in particular, Bola-DRB3) providing total immunity of cattle and specific resistance to certain diseases (8, 9).
The fourth unit includes monitoring of gene and chromosomal mutations. The dominance of several commercial breeds of domesticated species restricts their intra-breed variation and results in a rapid expansion of mutations originated in particular highly productive animals. The global translocation of gene pools of commercial species (eg., cattle) and their regional introduction necessitate the constant monitoring of such mutations, as BLAD (leukocyte adhesion deficiency), Mulefoot (mule's hoof), Dumps (deficit of uridinemonophosphate synthase), CVM (complex vertebral malformation), citrullinaemia and others; this also requires the screening of classical Robertsonian translocations (eg, 1/29) related to reproductive function and viability. At the same time, there are the gene mutations for myostatin - nt821 (del11) providing enforced formation of muscle mass (the phenomenon of “double muscle”), as well as other mutations correlated with accumulation of muscle mass in sheep and cattle, such as the autosomal mutation CLPG (calli pyge – “beautiful buttocks”).
The fifth unit (monitoring of veterinary and zootechnical parameters) implies organization of the system including observation of diseases, analysis of morphometric characteristics (exterior), commercially valuable traits and their dynamics, the size of animal population and quality of livestock. In this case, the linear methods of estimation of sires upon the quality of their progeny can be used to obtain non-biased assessments and increase the rate of genetic improvement of breeds’ commercially valuable properties (BLUP-method) (10). The main purpose of zootechnical monitoring are selection and preservation in generations of unique productivity indices of certain breeds. The veterinary monitoring aimed at diagnostics and test-identification of individuals resistant to epizooties, as well as revealing the reasons of local breeds’ resistance to some diseases.
The sixth unit accumulates the collected data and combines the elements of monitoring and control system for preservation in situ of domesticated animals. Modern mathematical methods and computer software provide the processing of large amount of data and fast calculation of many important parameters such as inbreeding coefficient and effective horizontal area of population. These methods are also used in cluster analysis, to detect violations in genetic equilibrium, genetic distances and divergence, to evaluate interactions between gene pools within and between breeds – this all makes mathematical and computer tests the integral part of monitoring system.
The seventh unit - planning (or simulation) of breeding work and selection of its optimal strategy. The data of overall observation of a studied population (breed) are the basis for sound decisions about the status of certain individuals or breeds, and these data are also used to develop science-supported programs for preservation of qualitative diversity upon the “genetic value” of the particular individual, population or a breed.
Pedigree farms play important role in genetic monitoring. The system of genefund farms is being formed in Russian Federation today – there are nearly 80 farm enterprises having a federal status of the genetic fund. Though, this is not enough. In the authors’ opinion, there should be from 600 to 1000 pedigree farms necessary for preservation of domestic genetic resources in livestock breeding. This calculation is based on data from the State Register of Breeding Achievements of the RF: by January 1, 2008, it included 33 198 breeds of domesticated species. The full maintenance and propagation of a breed requires three to five farms working along a single system of preservation and breeding plan with unified, constantly and effectively used methods of genetic monitoring. The gene pool of domestic breeds is an important resource for ongoing and future breeding work, and this is the factor closely linked with genetic and food security.
The proposed concept of genetic monitoring of domesticated animal breeds accumulates major genetic tools of contemporary science providing identification of genetic anomalies, as well as numerous genetic markers with different functions. It ensures great prospects for development of marker-assisted selection (MAS) in animal husbandry, when the breeder can select individuals by their genotype upon the tests of DNA molecules, their polymorphism, genes and loci, along with selection of animals by phenotype. Genome sequencing of major domesticated species makes possible the development of the relatively new branch of genetic science – genomics, which studies the genome, individual genes and their expression.
Thus, the scheme for monitoring of gene pools of domesticated animal species has been proposed, which makes possible to obtain the data on genotypic and phenotypic diversity, its assessment and analysis, and it also allows the creation of databases of breeds (herds) and species. The complex of contemporary science methods provides information necessary for optimization of breeding processes including preservation of gene pools. Genetic monitoring allows a substantiated choice and the “initial assessment”, and then it helps to determine the optimum and limits of acceptable changes. The data about structural genes, multi-loci spectra of DNA amplicons, mutational variability and other genomic characteristics of rare and endangered breeds may be used as indispensable sources of information for gene-geographic fundamental researches, for development of genetic strategies in programs for preservation of breeds, and for breeding animals with desired traits.
REFERENCES
1. Timofeef-Resovskii N.V. and Jablokov A.V., Phenes, Phenetics and Evolution Biology, Priroda, 1973, no. 5, pp. 40-51.
2. Fenetika prirodnykh populjatsij (Phenetics of Natural Populations), Ed. Jablokov A.V., Moscow, 1988.
3. Maksimenko V.F., Lobkov V.Yu., Zinov’eva N.A. and Sulimova G.E., Sistema geneticheskogo monitoringa jaroslavskoi porody krupnogo rogatogo skota (Gene Monitoring System in Cattle the Yaroslavl’ Breed), Yaroslavl’, 2005.
4. Glazko V.I., Stolpovskii Yu.A., Tarasuyk S.I., Bukarov N.G. and Popov N.A., The Genetic Structure of the Pintsgau Cattle Breed in the Carpathian Region, Genetika, 1996, vol. 32, no. 5, pp. 676-684.
5. Glazko V.I., Stolpovskii Yu.A., Duman’ T.N. and Kushnir A.V., Spatial and Temporal Distribution of Genetic and Phenetic Structure in the Grey Ukrainian Cattle, Dokl. NAN Ukrainy, 2000, no. 7, pp. 183-187.
6. Sulimova G.E., DNA Restriction Fragments Length Polymorphism in Farm Animals: Methods, Findings and Prospects, Usp. sovr. genetiki, 1993, vol. 18, pp. 3-35.
7. Stolpovskii Yu.A., Shmiit L.V., Kol N.V., Evsyukov A.N., Rusina M.N. and Churgui-ool O.I., Analysis of Genetic Variability and Phylogenetic Relations in Populations of Tuvinian Short-Fat-Tailed Seep with ISSR-Markers Usage, S.-kh. biol., 2009, no. 6, pp. 34-43.
8. Sulimova G.E., Udina I.G., Shaikhaev G.O. and Zakharov I.A., DNA-Polymorphism of the Cattle Gene BolA-DRB3 in Connection with Resistance and Susceptibility to Leucosis, Genetika, 1995, vol. 31, no. 9, pp. 1294-1299.
9. Moiseeva I.G., Ukhanov S.V., Stolpovskii Yu.A. et al., Genofondy sel'skokhozyaistvennykh zhivotnykh: geneticheskie resursy zhivotnovodstva Rossii (Genofunds of Farm Animals: Genetic Resources for Livestock Husbandry in Russia), Zakharov I.A., Ed.,Moscow, 2006.
10. Katkov L.V., Samorukov Yu.V., Chubar’ N.M. et al., Programma sokhraneniya i ratsional’nogo ispol’zovaniya genofonda krasnoi gorbatovskoi porody KRS (The Program for Preservation and Management of the Red Gorbatovskaya Cattle Breed), Dubrovitsy, 2001.
N.I. Vavilov Institute of General Genetics RAS, Moscow 119991, Russia e-mail: stolpovsky@vigg.ru |
February 15, 2010 |