ÓÄÊ 638.12:631.523:577.212:57.088
IDENTIFICATION OF RACE AND POPULATIONS OF HONEY-BEE WITH THE USE OF PCR-METHOD
N.I. Krivtsov1, I.I. Goryacheva2, I.G. Udina2, A.V. Borodachev1, M.A. Monakhova3
For the first time the authors made the molecular-biological investigation of genotypic structure in collection honey-bee colonies from pedigree apiaries. The high degree of purebred investigated honey-bee on mother line was revealed. The bee-families were determined, which can be used for a development of criteria of genetic passports of Central Russian breed of honey-bee.
The need in intense breeding work on honey-bee and conservation of its biological diversity are the important reasons for development of modern approaches to study and mobilization of available genetic resources. The complex study of prospects for using honey-bee colonies in breeding work can be performed upon the analysis of their morphometric and molecular genetic characteristics. A classical morphometric analysis allows determination of honey-bee breeds, which along with molecular and genetic properties helps to evaluate the characteristics of purebred honey-bee colonies (primarily, their queens).
The Central Russian breed of domestic honey-bee is uniquely adapted to rigorous local climate while having a number of commercially valuable traits unusual for other breeds - winter hardiness, high egg-laying qualities of bee queens and resistance to several diseases. This is the basic breed involved in breeding work on honey-bee in Central and Northern Russia, therefore, the assessment of breeding purity of honey-bee families in pedigree apiaries is extremely important. Such assessment of honey-bee on maternal line can be performed by investigation of marker region in the mitochondrial genome of Apis mellifera - the AT-rich intergenic locus located between the genes for cytochrome oxidase subunits I and II (COI-COII). In Central Russian breed, this region is formed by 3'-end of the gene for cytochrome oxidase I – the gene for tRNKLeu - P-element - two Q-elements -5'-end of the gene for cytochrome oxidase II. P-element has a length of 54 bp, it is fully composed by the nucleotides A + T. Q element - 196 bp length, contains 93,4% A + T. Total length of the locus is about 600 bp (1). In southern breeds of honey-bee (Mountain Grey Caucasian, Yellow Caucasian, Carpathian), this intergenic locus of nearly 300 bp length consists of only Q-element. Earlier, the structure of the intergenic locus COI-COII was analyzed in Southern Ural populations of Bashkir wild honey-bee and apiary honey-bee (2, 3).
The purpose of this work was to study breed purity of honey-bee families in breeding apiaries for evaluation the prospects for using these families in development of genetic criteria for certification of the Central Russian breed of honey-bee.
Technique. The object of study - Apis mellifera L. the Central Russian breed population of Orel Experimental Station for Beekeeping of Research and Development Institute of Beekeeping, as well as the families from populations of Orel, Kemerovo, Vologda, Perm’, Kirov and Krasnoyarsk provinces, from Bashkortostan, Tatarstan, Mordovia, Mari El and Altai. For comparison, the Maikop breed type of the Carpathian breed of honey-bee was analyzed.
DNA was prepared using a commercial kit for DNA extraction (“Isogene”, Russia) according to manufacturer’s instruction. To perform polymerase chain reaction (PCR), the original primers specific to, respectively, 3'-end of COI gene and to 5'-end of COII gene, were designed. PCR was performed using Encyclo PCR kit for amplification (“Evrogen”, Russia) under the following regime: initial denaturation - 95 °C, 2 min; 35 cycles including denaturation (95 °C, 15 s), annealing of primers (58 °C, 15 s), and polymerization (72 °C, 40s); the final polymerization - 72 °C, 4 min. Electrophoresis of amplification products was performed in 1% agarose gel. Before sequencing, PCR products were purified using JetQuiq Elution Kit (“Genomed”, Germany) following the manufacturer’s recommendations. Sequencing with both primers was carried out in the automated sequencer Applied Biosystems (USA).
Results. Central Russian honey-bee breed is spread on a large territory as a number of local populations. The analysis of exterior traits is widely used to determine breed purity of various breeds, but this method can’t reveal populations within a breed (Table). Most of differences in traditional exterior traits (morphological measurements) are unreliable and correspond to seasonal changes, so this criteria isn’t suitable for identification of local populations and it can be only used for differentiation of subspecies (breeds), particularly, Central Russian and Carpathian honey-bee.
Exterior traits of honey-bee Apis mellifera L. different breeds and populations (Õ±õ) |
|||
Breed, population |
Length of proboscis, mm |
Distance between peaks of the 3rd tergum, mm |
Cubital index, % |
Central Russian |
|
|
|
Bashkir (Kugarchi) |
6,34±0,024 |
4,86±0,022 |
54,7±1,37 |
Bashkir (Lemezy) |
6,20±0,060 |
5,06±0,019 |
59,3±1,28 |
Bashkir (central) |
6,21±0,024 |
4,90±0,017 |
60,8±1,36 |
Vologda |
6,17±0,035 |
5,02±0,020 |
65,0±1,27 |
Altai |
6,28±0,021 |
4,97±0,023 |
63,8±1,44 |
Kemerovo |
6,32±0,019 |
4,99±0,022 |
54,0±1,35 |
Kirov |
6,30±0,027 |
4,96±0,016 |
59,6±1,52 |
Krasnoyarsk |
6,15±0,036 |
5,12±0,018 |
65,0±1,36 |
Mari El |
6,22±0,018 |
4,94±0,024 |
61,5±1,34 |
Mordovia |
6,32±0,027 |
4,95±0,012 |
60,8±1,51 |
Orel |
6,29±0,057 |
5,12±0,023 |
60,8±1,72 |
Perm’ |
6,29±0,021 |
4,92±0,026 |
61,3±1,31 |
Tatarstan |
6,24±0,024 |
5,00±0,023 |
61,5±1,25 |
Carpathian, the Maikop breed type |
6,70±0,020 |
4,90±0,010 |
47,9±1,20 |
At the same time, there are the prospects for using molecular and genetic techniques for identification of local populations, since results of these tests are less dependent on environmental conditions and provide more accurate assessment of taxonomic location of the object.
After the amplification, PCR products were obtained for all the studied samples. When analyzing the DNA of Central Russian breed, the fragments of not less than 600 bp length were detected, which clearly indicates a breed purity on maternal line of the studied bee families. The structure of the studied fragment allowed to reveal genetic differentiation of breeds. In 30% families from population of Vologda and Perm’ provinces, Tatarstan and Bashkortostan, the longer fragments (800 bp and 1000 bp) of mitochondrial DNA (mtDNA) the intergenic locus COI-COII were found (Figure). The first of these “heavy” versions of the mitochondrial haplotypes is known to be described in literature: its structure PQQ is formed by repeating Q-element (1). Have performed sequencing of amplification products 600 bp length and their comparison with the database GenBank (EF676104), no mutations in the studied sequences were observed. As expected, the fragment of about 300 bp length was found in all honey-bees the Maikop type of Carpathian breed.
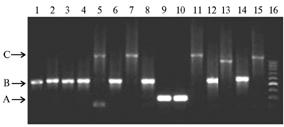 |
Agarose gel electrophoresis of amplification products of the intergenic locus COI-COII mtDNA in Central Russian breed local populations of honey-bee: samples 1-8 - Tatarstan; 9-10 - Kemerovo province; 11-12 - Vologda province, 13-15 - Orel province; 16 - molecular weight marker M100. Arrows show location of expected PCR fragments: A - 300 bp, B - 600 bp, C - 1000 bp. |
The analysis of length polymorphism of the mtDNA intergenic locus COI-COII in 10% bee families from pedigree apiaries of Orel Experimental Station confirmed their breed purity; no mitochondrial variants of the marker locus specific to southern breeds were revealed.
Studies of mitochondrial DNA of A. mellifera were started about 20 years ago (1). Currently, marker regions of mitochondrial genome are used to assesses the level of diversity in commercial populations of honey-bee (4), to clarify biogeography of subspecies (5), to establish genetic structure of populations (6, 7), to study hybridization of subspecies and introduction of bee species (8-10) . In some cases, a simultaneous analysis of peculiarities of mitochondrial DNA and microsatellites is carried out (4).
Identification of the structure of COI-COII is the first and necessary stage for development the criteria for genetic certification of honey-bee species in Russia. Genetic certification is of particular importance today owing to the main current direction of beekeeping - creation and maintenance of maternal and paternal lines, interlinear hybridization using instrumental insemination and selection of effective combinations among interlinear hybrids. Development of genetic passports of breeds and individual certification of most valuable parents is the way to increase efficiency of breeding work in beekeeping.
Thus, the study of genetic structure of the collection bee families from breeding apiaries was performed using PCR, which revealed a high degree of purebred bees on maternal line. The honey-bee colonies suitable for development of criteria for genetic certification of Central Russian breed have been determined.
REFERENCES
1. Cornuet J.M., Garnery L. and Solignac M., Putative Origin and Function of the Intergenic Region between COI and COII of Apis mellifera L. Mitochondrial DNA, Genetics, 1991, vol. 128, no. 2, pp. 393-403.
2. Nikonorov Yu.M., Ben’kovskaya G.V., Poskryakov A.V. et al., The Use of PCR Technique for Control of Pure-Breeding of Honeybee (Apis mellifera mellifera L.) Colonies from the Southern Urals, Genetika, 1998, vol. 34, no. 11, pp. 1574-1577.
3. Nikolenko A.G. and Poskryakov A.V., Polymorphism of Locus COI-COII of Mitochondrial DNA in the Honeybee Apis mellifera L. from the Southern Ural Region, Genetika, 2002, vol. 38, no. 4, pp. 458-462.
4. Il’yasov R.A., Petukhov A.V., Poskryakov A.V. and Nikolenko A.G., Local Honeybee (Apis mellifera mellifera L.) in the Urals, Genetika, 2007, vol. 43, no. 6, pp. 855-858.
5. Chapman N.C., Lim J. and Oldroyd B.P., Population Genetics of Commercial and Feral Honey Bees in Western Australia, J. Econ. Entomol., 2008, vol. 101, pp. 272-277.
6. Stevanovic J., Stanimirovic Z., Radakovic M. and Kovacevic S.R., Biogeographic Study of the Honey Bee (Apis mellifera L.) from Serbia, Bosnia and Herzegovina and Republic of Macedonia Based on Mitochondrial DNA Analyses, Genetika, 2010, vol. 46, pp. 685-691 (http://www.ncbi.nlm.nih.gov/pubmed/20583605).
7. De la Rua P., Galian J., Serrano J. and Moritz R.F., Genetic Structure and Distinctness of Apis mellifera L. Populations from the Canary Islands, Mol. Ecol., 2001, vol. 10, no. 7, pp. 1733-1742.
8. Palmer M.R., Smith D.R. and Kaftanoglu O., Turkish Honeybee: Genetic Variation and Evidence for a Fourth Lineage of Apis mellifera mtDNA, J. Heredity, 2000, vol. 91, pp. 42-46.
9. Franck P., Garnery L., Celebrano G., Solignac M. and Cornuet J.-M., Hybrid Origin of Honeybees from Italy (Apis mellifera ligustica) and Sicily (A. m. sicula), Mol. Ecol., 2000, vol. 9, no. 7, pp. 907-921.
10. Kraus F.B., Franck P. and Vandame R., Asymmetric Introgression of African Genes in Honeybee Populations (Apis mellifera L.) in Central Mexico, Heredity, 2007, vol. 99, pp. 233-240.
11. Collet T., Ferreira K.M., Arias M.C., Soares A.E. and Del Lama M.A., Genetic Structure of Africanized Honeybee Populations (Apis mellifera L.) from Brazil and Uruguay Viewed through Mitochondrial DNA COI-COII Patterns, Heredity, 2006, vol. 97, no. 5, pp. 329-335.
1All-Russia Research and Development Institute of Bee Keeping, RAAS, Ryazan’ province, Rybnoe 391110, Russia; |
Received November 25, 2009
|













