doi: 10.15389/agrobiology.2012.4.36eng
УДК 636.03:636.018:575.174.015.3
HOMOLOGOUS NUCLEOTIDE SEQUENCES OF THE FLANK OF RETROTRANSPOZON PawS5 OF R173 FAMILY IN ANIMAL AND PLANT GENOMES
V.I. Glazko, M.A. El’kina, T.T. Glazko
A comparative analysis was carried out of the amplification product spectra received in polymerase chain reaction with the use of both animal (cattle, sheep and horse breeds) and plant (rice varieties) DNA and the flank of retrotransposon PawS5 of R173 family as a primer. The breed-specific traits in animals and the variety-specific traits in rice were revealed in the amplicons spectra. The possible reasons for presence in genomes of animals and plants of the homologous nucleotide sequences associated with the retrotransposon, their polymorphism and efficiency of use as an anchor for a polylocus genomic genotyping of plants and animals are discussed.
Keywords: the inverted terminal retrotransposon repeats, IRAP-PCR markers, polylocus genotyping, polymorphic information contents, share of polymorphic loci.
Structural genomics and its implemented techniques has allowed the progress of applied population genetics from genotyping of single genes to entire genomes, from genetic description of population structure for several genes to multi-locus comparison of genotypes (from tens of genes to their complete sequencing) (1). Such polylocus genotyping was called “genome scanning” (2). This technique often implies the anchor function of mobile genetic elements (MGE) owing to their large number and high polymorphism in genomes (3). Some of MGE are very common and fragments homologous to MGE were recorded in distant taxa, eg. different classes of vertebrates (4), plants and animals (5), as well as in other taxa (6).
Genomic scanning is of particular interest for monitoring the dynamics of gene pools of agricultural species, which could help to develop genetically-based programs for preservation of endangered local animal breeds and crop varieties along with its use in conventional selection. According to FAO (Food and Agriculture Organization, http://www.fao.org), degradation of fertile soils, water resources, reduced biodiversity of breeds and varieties in agriculture are becoming rampant and actually incompatible with its sustainable development (7).
R173 family of retrotransposons was originally discovered in rye. This group of about 15 thousand individual copies per diploid genome is known to be spread over all chromosomes; such elements have the size ranging from 3 to 6 kbp, and two flanking regions highly homologous to flanks of the certain wheat retrotransposon (8-10). The system of primers designed upon R173 was found to be efficient in differentiation of closely related wild and cultivated plant species, varieties of potatoes and sunflower (11).
This work represents the authors’ attempt to assess the possibility of obtaining a universal anchor for genome-wide scanning, which was done though the comparative analysis of multi-locus spectra of DNA of major farm mammalian species (cattle, sheep, horses), and monocots (rice) using the terminal repeat of PawS5 retrotransposon of R173 family.
Technique. The objects of study were different livestock species and breeds - cattle (Black-and-White, Red Estonian and Yakut breeds), horses (Russian Trotter, Karachay and Altai breeds), sheep (Edilbaev, Kalmyk and Karachay breeds) (total 140 head) as well as 14 varieties of rice (collection of the All-Russia Research and Development Institute of Rice, Krasnodar) with different duration of growing season (early-ripening: Dal’nevostochny, Izumrud, Novator, Sadko, Sprint, Fontan, Fakel; mid-ripening: Viola, Liman, Primorsky, Rapan, Yantar’; late-ripening: Lider, Snezhinka).
To assess the polymorphism of DNA fragments flanked by inverted regions of PawS5 (AAC GAG GGG TTC GAG GCC) (Inter-Retrotransposon Amplified Polymorphism - IRAP) (8-10), genomic DNA was isolated from blood cells of animals and cells of seedlings using a commercial kit “DNA Extras-1” (“Synthol”, Russia). PCR was performed on a thermocycler “Tertsik” (Russia) using RT-PCR mixture kit (“Synthol”, Russia) under the following regime: initial elongation at 94 °С — 2 min, denaturation 94 °С — 30 s, annealing 55 °С — 30 s, elongation 72 °С — 2 min, final elongation 72 °С — 10 min (35 cycles). Amplification products were separated by electrophoresis in 1,5% agarose gel; a molecular weight marker DNA GeneRulerTM 100 bp DNA Ladder Plus (“MBI Fermentas”, USA) was used to determine the length of resulting fragments. The results were visualized by ethidium bromide staining, examined in UV light, and photographed.
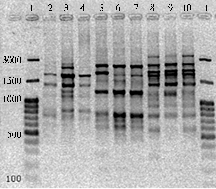 |
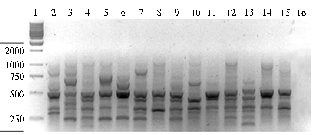
Fig. 1. Electrophoretic spectra of amplicons obtained through IRAP-PCR (Inter-Retrotransposon Amplified Polymorphism PCR) with terminal region of retrotransposon PawS5 used as a primer, in different animal species: 1, 11 — molecular weight marker GeneRulerTM 100 bp DNA Ladder Plus (“MBI Fermentas”, USA); 2, 3, 4 — cattle; 5, 6, 7 — sheep; 8, 9, 10 — horses (one individual of each breed). |
Computation of data was performed in a program TFPGA. Polymorphic information content (PIC) was determined using a formula for diallel loci: PIC = 2f(1 - f), where f - frequency of one of the two alleles. PIC mean was found over the entire spectrum of amplification products (PICm). The estimates of amplicons’ polymorphism were used to calculate genetic distances (DN, M. Nei, 1978) between groups, which values were used to draw a dendrogram. The sequences homologous to PawS5 were sought in Gen-Bank database of NCBI (National Center for Biotechnology Information) using a computer program BLASTn.
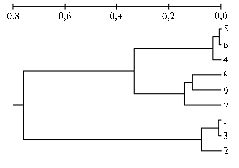
Results. Comparative analysis of genetic structure of farm animals populations (cattle, sheep, horses) performed upon the estimates of polymorphism of 25 DNA fragments of different lengths (Fig. 1) has revealed in the spectra of amplicons a number of DNA fragments ranging in size from 1,300 to 480 bp. Maximum values of PICm and proportion of polymorphic loci (Table 1) were found in local breeds of all three species compared to those in population largely controlled by artificial selection (Black-and-White cattle, racing trotters, Edilbaev sheep). Figure 2 shows the dendrogram of genetic relationships between the studied groups of animals plotted with the use of calculated genetic distances (DN) between genotypes upon the distribution of DNA fragments of different lengths flanked by inverted thermal repeat of PawS5.
The dendrogram includes two main clusters: the first one shows two sub-clusters (cattle and sheep), the second corresponds to horse breeds. These findings suggest that multi-locus genotyping using the terminal repeat of PawS5 as an anchor allows to identify actual events of genetic differentiation of animal species that can be clustered according to their known taxonomic relationships. In this work, each species was identified as one group of studied breeds, cattle and sheep belong to one cluster (even-toed ungulates), while the horse breeds form another independent cluster (odd-toed ungulates) (Fig. 2)
| 1. Characteristics of polymorphic amplification spectra obtained through IRAP-PCR (Inter-Retrotransposon Amplified Polymorphism PCR) with terminal repeat of retrotransposon PawS5 used as a primer, in different animal species | ||
Breed |
Proportion of polymorphic loci, % |
PIC |
Horses |
||
Altai |
20 |
0,096 |
Russian Trotter |
13 |
0,049 |
Karachay |
15 |
0,108 |
Sheep |
||
Karachay |
50 |
0,195 |
Edilbaev |
33 |
0,110 |
Kalmyk |
47 |
0,199 |
Cattle |
||
Black-and-White |
33 |
0,108 |
Yakut |
47 |
0,214 |
Red Estonian |
47 |
0,178 |
Note. PIC — Polymorphic information content. |
||
|
Fig. 2. Dendrogram of genetic distances (DN) between different animal species plotted upon the frequency of 25 loci identified in amplification spectra through IRAP-PCR (Inter-Retrotransposon Amplified Polymorphism) with terminal repeat of retrotransposon PawS5 used as a primer. Breeds of farm animals: 1 — Altai horse, 2 — Russian Trotter horse, 3 — Karachay horse, 4 — Karachay sheep, 5 — Edilbaev sheep, 6 — Kalmyk sheep, 7 — Black-and-White cattle, 8 — Yakut cattle, 9 — Red Estonian cattle. |
IRAP-PCR of rice varieties (Fig. 3) has provided a similar spectrum of amplicons which included 13 major fragments of 1000 –200 bp lengths clearly reproducible in repeated amplifications and considered as major amplicons of the spectrum; DNA fragments difficult for precise identification (minor amplicons) were not accounted in repeated amplifications. PCR with the abovementioned primer revealed the absence of any intravarietal differences for major amplicons, while intervarietal differences were recorded: the number of amplicons ranged from 5 (cv Sadko, Fontan, Yantar’) to 9 (cv Dal’nevostochny, Fakel, Snezhinka).
In amplification spectra of almost all studied varieties there were regularly present fragments of 500, 450, 350 and 250 bp length (Table 2), which were significantly shorter than the retrotransposon (from 3 to 6 kbp) (3, 4). This fact allows assuming that in the genomes of different rice varieties the terminal regions of PawS5 maintain relatively constant reciprocal proximity (250-500 bp) allowing alternate DNA chains.
|
Fig. 3. Electrophoretic spectra of amplicons obtained through IRAP-PCR (Inter-Retrotransposon Amplified Polymorphism PCR) wigh terminal repeat of retrotransposon PawS5 used as a primer, in different rice varieties: 1, 11 — molecular weight marker GeneRulerTM 100 bp DNA Ladder Plus (“MBI Fermentas”, USA); 2 —Viola, 3 — Dal‘nevostochny, 4 — Izumrud, 5 — Lider, 6 — Liman, 7 — Novator, 8 — Primorsky, 9 — Rapan, 10 — Sadko, 11 — Sprint, 12 — Snezhinka, 13 — Fakel, 14 — Fontan15 — Yantar’, 16 — negative control. |
2. Characteristics of polymorphic spectra of amplicons obtained through IRAP-PCR (Inter-Retrotransposon Amplified Polymorphism PCR) with terminal repeat of retrotransposon PawS5 used as a primer, in different rice varieties |
||||||||||||||
Variety |
Amplicon length, bp |
Number of amplicons |
||||||||||||
1000 |
950 |
850 |
750 |
700 |
600 |
500 |
450 |
400 |
350 |
300 |
250 |
200 |
||
Dal’nevostochny |
+ |
0 |
0 |
+ |
+ |
0 |
+ |
+ |
+ |
+ |
+ |
+ |
0 |
9 |
Izumrud |
+ |
0 |
0 |
+ |
0 |
0 |
+ |
+ |
0 |
+ |
0 |
+ |
+ |
7 |
Novator |
+ |
0 |
+ |
0 |
0 |
0 |
+ |
+ |
0 |
+ |
+ |
+ |
+ |
8 |
Sadko |
+ |
0 |
0 |
0 |
+ |
0 |
0 |
+ |
0 |
+ |
+ |
0 |
0 |
5 |
Sprint |
0 |
0 |
0 |
0 |
+ |
0 |
+ |
+ |
+ |
+ |
+ |
0 |
0 |
6 |
Fakel |
0 |
0 |
0 |
+ |
+ |
0 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
9 |
Fontan |
+ |
+ |
0 |
0 |
0 |
0 |
+ |
+ |
0 |
+ |
0 |
0 |
0 |
5 |
Viola |
0 |
0 |
0 |
+ |
0 |
+ |
+ |
+ |
0 |
+ |
+ |
0 |
0 |
6 |
Liman |
0 |
0 |
0 |
0 |
+ |
+ |
+ |
+ |
+ |
+ |
0 |
+ |
0 |
7 |
Primorsky |
+ |
0 |
+ |
0 |
0 |
0 |
+ |
+ |
0 |
+ |
0 |
+ |
0 |
6 |
Yantar’ |
0 |
0 |
+ |
0 |
0 |
0 |
+ |
+ |
0 |
+ |
0 |
+ |
0 |
5 |
Lider |
+ |
0 |
0 |
0 |
+ |
0 |
+ |
+ |
+ |
+ |
0 |
+ |
0 |
7 |
Rapan |
0 |
0 |
0 |
0 |
0 |
+ |
+ |
+ |
0 |
+ |
+ |
+ |
0 |
6 |
Snezhinka |
+ |
0 |
0 |
+ |
0 |
0 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
9 |
The diversity of amplification products indicates that insertions / deletions of such fragments occur fairly frequently in the studied rice varieties. Fast evolution of the rice genome associated with transposable elements was described in detail in the literature (12): in this species the frequency of deletions and insertions of transposable elements twice exceeds the frequency of synonymous substitutions on the average for structural genes. In wheat, increased mobility of R173 elements provided intravarietal diversity of their presence / absence associated with storage life of seeds (13). Apparently, such rapid evolution of transposing sequences in the genome of rice explains the unique combination of DNA amplicons flanked by inverted repeat of retrotransposon PawS5 peculiar to each of the 14 studied species.
Interestingly, that the frequency of short (less than 480 bp) major DNA fragments flanked by inverted terminal repeats of PawS5 was higher in the studied genomes of rice varieties compared with the mammalians’ genomes (Fig. 1, 2). Perhaps, this fact was concerned with smaller size of rice genome (nearly 6 times shorter) than that of even- and odd-toed ungulates.
Searching in GenBank for sequences homologous to PawS5 fragment there was found a large number of fragments with partial homology. In mammals, as a rule, they were related to a polygenic family P450 and genes associated with the immune system, as well as transcription regulatory factors. In general, the regions homologous to the flank of PawS5 were found in a wide taxonomic area including prokaryotes.
In recent years, a widespread horizontal transfer of various mobile genetic elements has been reported in various taxa, such as Helitron transpositions between fungi, plants and animals (5), Helitron transpositions between distant taxa of animals (6), retrotranspositions between crops (14). Findings of this research suggest that genomic relationships between different taxa through mobile genetic elements can be much more significant than what is traditionally considered.
Thus, homologous sequences present in the genomes of farm animals and cultivated crops can perform the role of anchor in genomic scanning and multi-locus genotyping. In this work, the terminal repeat of PawS5 was used a primer in polymerase chain reaction, which provided polylocus spectra of amplification products allowing precise differentiation of both animal breeds (cattle, sheep and horses) and crop varieties (rice).
REFERENCES
1. Luikart G., England P.R., Tallmon D. et al., The Power and Promise of Population Genomics: from Genotyping to Genome Typing, Nat. Rev. Genet., 2003, vol. 4, pp. 981-994.
2. Manel S., Schwartz M.K., Luikart G. et al., Landscape Genetics: Combining Landscape Ecology and Population Genetics, Trends Ecol. Evol., 2003, vol. 18, pp. 189-197.
3. Mei L., Ding X., Tsang S.-Y. et al., AluScan: a Method for Genome-Wide Scanning of Sequence and Structure Variations in the Human Genome, BMC Genomics, 2011, vol. 12, pp. 564-573.
4. Kordis D. and Gubensek F., Unusual Horizontal Transfer of a Long Interspersed Nuclear Element between Distant Vertebrate Classes, PNAS USA, 1998, vol. 95, pp. 10704-10709.
5. Yang L. and Bennetzen J.L., Structure-Based Discovery and Description of Plant and Animal Helitrons, PNAS USA, 2009, vol. 106, pp. 12832-12837.
6. Thomas J., Schaack S., and Pritham E.J., Рervasive Horizontal Transfer of Rolling-Circle Transposons among Animals, Genome Biol. Evol., 2010, vol. 2, pp. 656-664.
7. The State of the World’s Animal Genetic Resources for Food and Agriculture, Rischkowsky B. and Pilling D., Eds., Rome: FAO, 2007.
8. Rogowsky P.M., Manning S., Liu J.-Y., and Langridge P., The R173 Family of Rye-Specific Repetitive DNA Sequences: a Structural Analysis, Genome, 1991, vol. 34, pp. 88-95.
9. Rogowsky P.M., Liu J.Y., Manning S., Taylor C., and Langridge P., Structural Heterogeneity in the R173 Family of Rye-Specific Repetitive DNA Sequences, Plant Mol. Biol., 1992, vol. 20, no. 1, pp. 95-102.
10. Rogowsky Р.М., Shepherd K.W., and Laпgridge Р., Polymerase Chain Reaction Based Mapping of Rye Involving Repeated DNA Sequences, Gеnоmе, 1992, vol. 35, no. 4, pp. 621-628.
11. Zaitsev V.S. and Khavkin E.E., Identification of Plant Genotypes by PCR Analysis of the R173 Family of Dispersed Repetitive Sequences, Dokl. RASKhN, 2001, no. 2, pp. 3-5.
12. Ма J. and Bennetzen J.L., Rapid Recent Growth and Divergence of Rice Nuclear Genomes, PNAS USA, 2004, vol. 101, no. 34, pp. 12404-12410.
13. Chwedorzewska K.J., Bednarek Р.Т. and Puchalski J., Studies on Changes in Specific Rye Genome Regions due to Seed Aging and Regeneration, Сеll Моl. Biol. Lett., vol. 2002, no. 7(2А), pp. 569-576.
14. Roulin А., Piegu B., Fortune P.M., Sabot F., D’Hont A., Manicacci D., and Panaud O., Whole Genome Surveys of Rice, Maize and Sorghum Reveal Multiple Horizontal Transfers of the LTR-Retrotransposon Route66 in Poaceae, BMC Evol. Biol., 2009, vol. 9, pp. 58 (http://www.biomedcentral.com/1471-2148/9/58).
K.A. Timiryazev Russian State Agrarian University, |
Received April 20, 2012
|