doi: 10.15389/agrobiology.2012.4.42eng
УДК 636.22/.28:636.082.12/.13:577
GENETIC CHARACTERISTIC OF GENEALOGICAL STRUCTURE OF KOSTROMSKAYA CATTLE BREED
S.G. Belokurov1, G.A. Badin2, O.S. Egorov2, A.V. Perchun2, G.E. Sulimova3
The present study investigated a genealogical structure of Kostromskaya cattle breed on blood groups in bulls-sires from the main farm bloodlines and related animal groups with different portion of Schwiss cattle blood used during the last years in breeding farms to improve milk productivity of Kostromskaya cattle. The spectrum of EAB-locus alleles of blood groups was analyzed and the genetic diversity, the degree of homozygosis and genetic consolidity of these genealogical groups were estimated. It was revealed, that Schwiss bulls-sires of American, Canadian and Austrian selection continue to influence, however, the studied farm lines and related groups have significant genetic distinguishes, confirmed by an index of genetic distance (DN = 0.647).
Keywords: genealogical structure, genetic diversity, related group, breeding line, blood groups, EAB-alleles, gene pool.
Genealogical structure of a particular breed or its structural units (lines, families, types, and related groups) is commonly used as an estimate of genetic diversity in livestock populations. This method is simple, but it has a significant flaw: many individuals originate from ancestors-representatives of different lines.
In recent studies, genealogical structure of breeds is often studied jointly with individual polymorphisms of proteins and blood groups. These substances remain unchanged in ontogeny, usually inherited in a codominant pattern and easy to determine by laboratory tests at early developmental stages. Such features of polymorphic proteins in biological fluids and blood groups were considered to use them as genetic markers in a wide range of theoretical and applied researches of animal husbandry: managed genealogical structure of breeds and their genetic consolidation, directed improvement of breeds, compatibility of different breeds in crosses aimed at heterosis and advanced breeding qualities of livestock, etc. Besides, immunogenetic techniques are a valuable tool in comparative studies of intrabreed livestock populations, because they reveal mechanisms underlying relative stability of structural units of a breed and its further development.
Inheritance of blood groups in cattle is controlled by allelomorphic genes located in 13 loci on different chromosomes. Accordingly, there are13 independent blood types in cattle, which systems are assumed as simple and complex. Simple blood groups are associated with one or two antigenic factors and two alternative alleles in corresponding loci. Complex systems of blood groups include two or more antigens usually responsible for several factors. The most complex B-system of blood types includes nearly half of all known antigens that determine over 400 blood groups. In such complex systems each blood group is controlled by a separate allele, and it is inherited as a whole from parents to offspring regardless of the number of antigens (1).
Kostroma cattle is one of the most valuable domestic breeds. This breed was established through complex reproductive crosses in farms of Kostroma and Nerekhta districts of Kostroma province. The foundation of Kostroma cattle was aboriginal cattle with gradual later involvement of Algau, Swiss and other foreign breeds. Kostroma breed is appreciated for high dairy and beef production along with unique biological properties. These animals are well-adapted to local conditions of feeding and keeping, they have a strong constitution, good health and resistance to bovine leucosis, and high dairy yield through a long productive life. Genetic potential of Kostroma cows allows producing more than 6000 kg milk annually (the best cows – up to 10-16 thousand kg) with high contents of milk fat (3,9-4,0 %) and protein (3,49-3,64 %) (2). High protein content in the milk and high quality composition of milk protein fractions are unique features of Kostroma breed. DNA testing of cows bred in the pedigree farm “Karavaevo” in respect to k-casein gene revealed 14 % AA homozygotes, 52 % AB heterozygotes, and 34 % cows with BB genotype desired for cheese-making quality of milk and increased yield of product (cottage cheese, hard and semi-hard cheese) from the unit of collected milk (3).
In recent years genealogical structure of Kostroma cattle was influenced by target breeding of dairy-oriented cows through crosses with Swiss Brown sires from the USA, Canada, Austria and Germany (4). At the same time, it’s important to improve Kostroma breed with a special role to breeding lines, which necessitates a well-working system of breeding, testing, selection and intense use of best bulls-sires (5).
The purpose of this work was studying and systematization of allelotypes of Kostroma bulls-sires used in pedigree farms of Kostroma province in recent years, in order to determine genetic distances between genealogical groups within the breed, and give recommendations for further use in breeding practice.
Technique. The data were obtained from 589 bulls-sires – representatives of current genealogical structure of Kostroma breed. A part of them were members of eight breeding lines: Caro KTKS-101 (n = 46), Ladok KTKS-253 (n = 74), Peek KTKS-419 (n = 15), Salat KTKS-83 (n = 10), Silach KTKS-84 (n = 5), Banan KTKS-333 (n = 17), Barkhat VDKS-6 (n = 4), and Ograd VDKS-24 (n = 4); the others were from eight related groups with different shares of Swiss Brown bloods: Kontsentrat 106 157 (n = 69), Meridian 90827 (n = 156), Master 106 902 (n = 133), Butler 107 506 (n = 23), Hill 76059 (n = 10), Keeper 55163 (n = 5) , Oregon 342 995 (n = 8), and Laird 71151 (n = 10).
The spectrum of EAB- alleles determining blood groups in cattle was studied by testing all animals for blood antigenic factors as described by P.F. Fortieth (6) with the use of standard immunospecific sera (including ones of own production), produced in the laboratory of immunogenetics of the Breeding Center (Association) for Red Cattle Breeds and confirmed by comparative trials. The tests were performed using 48-65 sera reagents for determination of antigens specific to ten systems of blood groups. Genetic characteristics of breeding lines and related groups were described by algorithms of genetic similarity (rm) according to K. Maijala and G. Lindstrom (7), and genetic distances (DN) according to M. Nei (8). The degree of homozygosity (Ca) was determined as a sum of squared frequencies of alleles of EAB-locus using the formula proposed by J. Matoushek (9). Dendrograms of immunogenetic distances were designed upon unweighted pairwise clustering of characteristics according to A. Masurov and V. Cherkaschenko (10). The studied alleles of bulls-sires were corrected upon the data of a reference manual (11).
Statistical calculations were performed in Microsoft Excel.
Results. The analysis of allelic pool of bulls-sires has revealed peculiarities of EAB alleles and their frequency patterns. In the whole studied group of bulls-sires there were revealed 32 alleles of EAB locus. Top frequent alleles: B1O3Y2A'2E'3G'P'Q'Y' (0,1474), B1G2KE'3F'2O' (0,0757), B2G3QT1A'1P' (0,0681), B2P1Y2G'Y' (0,0539), G3O1T1Y2E'3F'2 (0,1776), I1G'G" (0,1121), О' (0,0903), «b» (0,1818); total frequency of these alleles was 0,9069. The alleles specific to breeding lines: B1G2KE'1F'2O' (Salat KTKS-83), B2G3QT1A'1P' (Caro KTKS-101), I1G'G" and “b” (Ladok KTKS-253) with total frequency of 0,6713, to related groups — B1O3Y2A'2E'3G'P'Q'Y' (Meridian 90827), B2P1Y2G¢Y¢ and G3O1T1Y2E'3F'2 (Master 106902) with total frequency of 0,5289. Alleles B1O3Y2A'2E'3G'P'Q'Y' and “b” were found relatively frequent in breeding lines (respectively 0,1133 and 0,1874) as well as in related groups (0,1814 and 0,1761). At the same time, A'2G'G" (0,0564) and Y2A'1D'E'1 (0,0290) - labeling alleles in the related group Kontsentrat 106157 – were not found in studied breeding lines of Kostroma cattle.
The degree of homozygosity (Ca) over all tested bulls-sires averaged 19,54%; in related groups – 17,42 %, in breeding lines – 20,39 %. This high level of homozygosity resulted from the intense involvement of a limited number of best sires, whose sperm was used in crosses aimed at increased productivity (mainly Swiss breed and their offspring with varying degrees of pure blood), so it has led to elimination of certain allelic variants of EAB-locus. This fact increases the risk of reduced reproductive qualities and adaptivity of Kostroma cattle.
The index of genetic similarity (rm) between bulls-sires of major breeding lines and related groups averaged 0,510 (Table). The minimum similarity (0,059) was found in comparison of related group Laird 71151 and the line Barkhat VDKS-6, the maximum (0,921) - between related groups Meridian 90827 and Master 106902. Along with it, the comparison of related group Hill 76059 with the line Silach KTKS-84 showed similarity index 0,712, but it turned to 0,621 when the same group was compared with the lines Salat KTKS-83, Barkhat VDKS-6 and Ograd VDKS-24; this also proves accumulation of certain typical alleles after the use of a limited group of bulls-sires in order to obtain dairy type of cattle. Similar genetic effects were revealed by comparison of other breeding lines and related groups of Kostroma cattle.
Genetic resemblance (rm) and genetic distances (DN) between breeding lines and related groups of Kostroma cattle calculated upon the analysis of frequency of EAB-alleles in carriers bulls-sires (n = 589) |
||||||||||||||||
rm |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
1 |
|
0,644 |
0,538 |
0,450 |
0,584 |
0,759 |
0,626 |
0,722 |
0,345 |
0,376 |
0,259 |
0,430 |
0,428 |
0,202 |
0,280 |
0,194 |
2 |
0,356 |
|
0,406 |
0,419 |
0,577 |
0,472 |
0,434 |
0,535 |
0,392 |
0,403 |
0,469 |
0,452 |
0,284 |
0,422 |
0,472 |
0,440 |
3 |
0,462 |
0,594 |
|
0,654 |
0,601 |
0,626 |
0,595 |
0,384 |
0,335 |
0,319 |
0,444 |
0,418 |
0,262 |
0,484 |
0,457 |
0,401 |
4 |
0,550 |
0,581 |
0,346 |
|
0,622 |
0,443 |
0,344 |
0,476 |
0,346 |
0,295 |
0,318 |
0,481 |
0,350 |
0,299 |
0,327 |
0,168 |
5 |
0,416 |
0,423 |
0,399 |
0,378 |
|
0,580 |
0,428 |
0,559 |
0,429 |
0,335 |
0,466 |
0,406 |
0,285 |
0,621 |
0,406 |
0,299 |
6 |
0,241 |
0,528 |
0,374 |
0,557 |
0,420 |
|
0,693 |
0,504 |
0,408 |
0,347 |
0,570 |
0,352 |
0,208 |
0,712 |
0,611 |
0,686 |
7 |
0,374 |
0,566 |
0,405 |
0,656 |
0,572 |
0,307 |
|
0,500 |
0,227 |
0,155 |
0,399 |
0,316 |
0,059 |
0,621 |
0,441 |
0,583 |
8 |
0,278 |
0,465 |
0,616 |
0,524 |
0,441 |
0,496 |
0,500 |
|
0,595 |
0,436 |
0,573 |
0,449 |
0,295 |
0,621 |
0,378 |
0,583 |
9 |
0,655 |
0,608 |
0,665 |
0,654 |
0,571 |
0,592 |
0,773 |
0,405 |
|
0,921 |
0,882 |
0,762 |
0,727 |
0,779 |
0,726 |
0,712 |
10 |
0,624 |
0,597 |
0,681 |
0,705 |
0,665 |
0,653 |
0,845 |
0,564 |
0,079 |
|
0,879 |
0,774 |
0,772 |
0,740 |
0,812 |
0,724 |
11 |
0,741 |
0,531 |
0,556 |
0,682 |
0,534 |
0,430 |
0,601 |
0,427 |
0,118 |
0,121 |
|
0,720 |
0,617 |
0,850 |
0,837 |
0,828 |
12 |
0,570 |
0,548 |
0,582 |
0,519 |
0,594 |
0,648 |
0,684 |
0,551 |
0,238 |
0,226 |
0,280 |
|
0,598 |
0,593 |
0,675 |
0,536 |
13 |
0,572 |
0,716 |
0,738 |
0,650 |
0,715 |
0,792 |
0,941 |
0,705 |
0,273 |
0,228 |
0,383 |
0,402 |
|
0,565 |
0,547 |
0,442 |
14 |
0,798 |
0,578 |
0,516 |
0,701 |
0,379 |
0,288 |
0,379 |
0,379 |
0,221 |
0,260 |
0,150 |
0,407 |
0,435 |
|
0,822 |
0,913 |
15 |
0,720 |
0,528 |
0,543 |
0,673 |
0,594 |
0,389 |
0,559 |
0,622 |
0,274 |
0,188 |
0,163 |
0,325 |
0,453 |
0,178 |
|
0,812 |
16 |
0,806 |
0,560 |
0,599 |
0,832 |
0,701 |
0,314 |
0,417 |
0,417 |
0,288 |
0,276 |
0,172 |
0,464 |
0,558 |
0,087 |
0,188 |
|
Note. Breeding lines Ladok (1), Caro (2), Banan (3), Peek (4), Salat (5), Silach (6), Barkhat (7) and Ograd (8); related groups Meridian (9), Master (10), Kontsentrat (11), Butler (12), Laird (13), Hill (14), Keeper (15) and Oregon (16). |
||||||||||||||||
Antigenic composition of blood groups was illustrated by two clusters of breeding lines and related groups (Figure). The first cluster included bulls of related groups Kontsentrat 106157, Meridian 90827, Master 106902, Butler 107506, Hill 76059, Keeper 55163, Oregona 71151 and Laird 342995, the second - only breeding lines Sinach KTKS-84, Ladok KTKS -253, Caro KTKS-101, Pick KTKS-419, Salat KTKS-83, Banan KTKS-333, Ograd VDKS-24 and Barkhat VDKS-6.
Within the cluster of breeding lines, the minimum genetic distance have the lines Ladok KTKS-253 and Silach KTKS-84 (DN = 0,241), the maximum (DN = 0,514) - between the sub-cluster formed by the lines Salat KTKS-83, Banan KTKS-33, Pick KTKS-419 and another sub-cluster including the lines Caro KTKS-101, Ograd VDKS-24, Barkhat VDKS-6, Silach KTKS-84 and Ladok KTKS-253. In the cluster of related groups, the minimum genetic distance showed the groups Meridian 90827 and Master 106902 (DN = 0,079), the maximum (DN = 0,399) - the group Laird 71151 and a sub-cluster formed by Master 106 902, Meridian 90827, Kontsentrat 106157, Hill 76059, Oregon 342995, Keeper 55163, and Butler 107506.
Genetic distance (DN) equal to 0,647 between the first and second clusters of bulls-sires indicates genetic dissociation of these groups. Therefore, to obtain heterosis effects it is desirable to cross animals from the first cluster (related groups) with individuals from the second cluster (breeding lines).
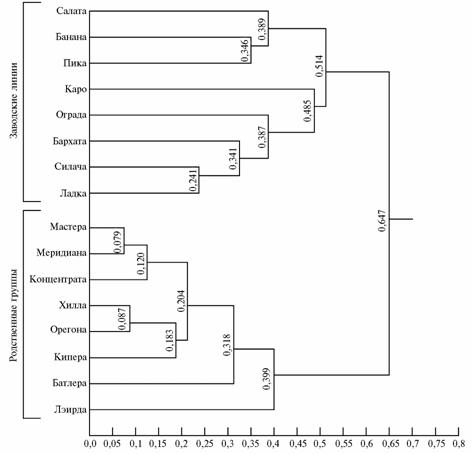 |
Dendrogram illustrating genetic relationships between major breeding lines and related groups of Kostroma cattle upon the analysis of frequencies of EAB-alleles in carriers bulls-sires with the use of non-weighted pairwise clustering (according to Mashurov A. and Cherkaschenko B.) (10). Numbers correspond to genetic distances calculated as described by М. Nei (8) (n = 589). |
Thus, even though a significant impact of Swiss Brown bulls-sires in past and present of Kostroma breed, gene pools of studied breeding lines and related groups have specific genetic differences. Genetic patterns of Kostroma cattle including peculiarities of blood types should be considered in breeding programs with pedigree cattle, as well as in common practice of selection of replacement livestock, target crosses, establishment of bulls sires, because these features are quite important for preservation and optimization of unique gene pool of Kostroma breed.
REFERENCES
1. Kolesnik N.N., Immunogeneticheskiesistemyvselektsiizhivotnykh(Immunogenetic Systems in Animal Breeding), Kyiv, 1972.
2. Lyagin F.F. and Badin G.A., Kostroma Breed of Cattle is Our Trademark, in 60 let kostromskoy porode krupnogo rogatogo skota: materially yubileynoi nauchno-prakticheskoi konferentsii (Papers of Sci.-Pract. Conf. “60th Anniversary of Kostroma Cattle Breed), Kostroma, 2004, pp. 58-67.
3. Bogdanova T.V., Badin G.A., Kalashnikova L.A. et. al., DNA-Based Assessment of Pedigree Kostroma Cattle, in Mat. 54-i Mezhvuz. nauch.-prakt. konf. “Aktual’nye problemy nauki v agropromyshlennom komplekse” (Papers of 54th Sci.-Pract. Conf. “Modern Issues of Science in Agriculture”), Kostroma, 2003, vol. 1, pp. 60-61.
4. Baranov A.V., Genetic Labeling and Its Use in Improvement of Dairy Cattle, Extended Abstract of Doctoral Sci. Dissertation, Moscow, 1997.
5. Shalugin B.V., Smirnova V.S., and Koroleva E.A., Genetic Potential of Kostroma Cattle Genealogical Groups Bred in Pedigree Farms and Its Realization, in Mat. 63-i Mezhvuz. nauch.-prakt. konf. “Aktual’nye problemy nauki v agropromyshlennom komplekse” (Papers of 63rd Sci.-Pract. Conf. “Modern Issues of Science in Agriculture”), Kostroma, 2012, vol. 1, pp. 149-153.
6. Sorokovoy P.F., Metodicheskie rekomendatsii po issledovaniyu i ispol’zovaniyu grupp krovi v selektsii krupnogo rogatogo skota (Guidelines on Determination of Blood Group in Cattle and Its Use in Breeding Practice), Dubrovitsy, 1974.
7. Maijala K. and Lindstrom G., Frequencies of Blood Group Genes and Factors in the Finnish Cattle Breeds with Special Regard to Breed Comparison, Ann. Agr. Fenn., 1966, vol. 5, pp. 76-93.
8. Nei M., Genetic Distance between Populations, Amer. Naturalist, 1972, vol. 106, pp. 283-292.
9. Matoushek J., Gruppy krovi u krupnogo rogatogo skota (Blood Groups of Cattle), Kyiv, 1964.
10. Mashurov A.M. and Cherkaschenko V.I., Considering Genetic Distances between Breeds during Selection, Zhivotnovodstvo, 1987, vol. 2, pp. 21-23.
11. Popov N.A. and Eskin G.V., Allelofond porod krupnogo rogatogo skota po EAB-lokusu. Spravochnyi katalog (Allelic Pool of Cattle Breeds for EAB-Locus: Reference Catalog), Moscow, 2000.
1Kostroma State Agricultural Academy, Karavaevo campus |
Received March 14, 2012
|









