doi: 10.15389/agrobiology.2012.4.69eng
УДК 636.2:619:616.155.392
A STUDY OF HETEROGENEITY OF BOVINE LEUKEMIA VIRUS GENOTYPES IN CATTLE
N.V. Bateneva, P.N. Smirnov, I.V. Mikhnovich
By polymerase chain reaction (PCR), a proviral DNA was tested in 762 blood samples from cattle of different breeds, sick and infected with bovine leukemia virus, to examine a viral heterogeneity. Using the Vector NTI software, the appropriate endonucleases were fond out for RFLP analysis. Based on data obtained from the analysis of polymorphisms of restriction fragment length, the 11 groups were defined as genotypes according to the types of patterns.
Keywords: bovine leukemia virus (BLV), genotypes, restriction enzyme, polymerase chain reaction, breeds of cattle, gag, p24.
Bovine leukemia virus, or BLV, is an infectious retrovirus. Three major structural genes encoding viral proteins of BLV are located in the following order: 5'-gag-pol-env-3 '. The gene gag encodes proteins (p24) that form capsid, they are necessary for the intracellular assembly and release from cells.
BLV is widespread in cattle over the world. Its genotypic diversity is an important issue of general biology and cattle breeding. At the same time, a limited number of published works on this item complicates any definite opinion on BLV (1-13) even though the data about the course of leukemic infection, its etiological agent (3, 5, 7), and high pathogenicity of certain BLV genotypes (2).
Most authors studied BLV in respect to its env-gene and only few of them – gag-gene (1, 2, 6, 7, 10). Variability of gag gene of BLV wasn’t reported in the available scientific literature. At the same time, gag-gene of BLV can be identified 5-7 months earlier than env-gene (3, 4).
This work presents the authors’ attempt to reveal genotypic variability of gag-gene of bovine leukemia virus (BLV).
Technique. The authors tested 762 blood samples obtained from cattle of different breeds bred in farm enterprises of Krasnodar krai (2006-2009) that were identified as seropositive in agar gel immunodiffusion test (AGID+) with a strain gp 51 BLV.
DNA of BLV provirus was isolated from blood leukocytes by sorbent assay with a set Diatom DNA Prep 100 (Isogen Lab. Ltd., Russia) according to the manufacturer's guidelines. PCR was performed using test kits GenePak ™ PCR test (Isogen Lab. Ltd., Russia) including positive and negative controls for BLV (gag).
PCR regime proposed by Mohammadabadi M.R. (10) was optimized in the view of expected fragment length. Elongation cycles were selected empirically for greatest yield of the specific amplicon while a minimum content of nonspecific fragments. The reaction was carried out in the thermocycler Master-cycler Gradient (Eppendorf AG, Germany). The reaction volume - 40 ml. PCR regime: initial denaturation – 94 °С for 3 min; 42 cycles (denaturation – 94 °С, 15 s; annealing – 58 °С, 15 s; elongation – 72 °С, 30 s); final elongation – 72 °С, 3 min.
The carriers of BLV gag gene (p24) (expected fragment length 347 bp) were identified using oligonucleotide primers 5'-GGA GGW GGR AAG ATG CGA ACT ATT-3' and 5'-GTC CGY TCT ACY AAC CCT GAA CTT-3' (10) The primers were designed considering polymerase sequence of gag-gene full-size copy from the database of NSBI (National Center for Biotechnology Information, gi40796166, NP_777381.1) and synthesized in Medigen Lab. (Novosibirsk).
The structure of most variable regions was analyzed in Vector NTI program in order to select restriction endonucleases for the assessment of polymorphism.
Results. The length of gag-gene fragment flanked by the specific primers was 347 bp. Figure 1 shows a typical electrophoregram of amplification products with viral DNA integrated into the DNA of host animal. In all analyzed samples there was detected DNA fragment corresponding to the estimated length of amplicon (347 bp).
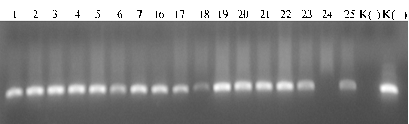 |
Fig.1. Typical electrophoregram of PCR-amplification products including BLV gag-gene proviral DNA fragment (347 bp) derived from DNA samples (1-25) of Red Steppe cattle (the farm enterprise “Kolos”, Krasnodar krai, Dinsky district): К(-) — negative control, К(+) — positive control (GenePak™ PCR test, Isogen Lab. Ltd., Moscow). |
PCR analysis was performed to reveal the presence of BLV proviral DNA in cattle using the primers specific for gag-gene region of 347 bp. Over all tested cattle the proportion of AGID-seropositive animals with negative PCR results was 2,6% (Table 1).
| 1. Occurrence of bovine leukemia virus the proviral DNA (gag gene fragment, 347 bp) detected by PCR in cattle individuals seropositive to agar gel immunodiffusion test (AGID) (Krasnodar krai, 2006-2009) | |||
Cattle breeds |
Number of individuals |
||
AGID+ |
AGID+, PCR+ |
AGID+, PCR- |
|
Black-and-White Holsteinized |
284 |
263 |
21 |
Red Steppe |
255 |
244 |
11 |
Ayrshire |
223 |
217 |
6 |
Total |
762 |
724 |
38 |
Note. Farm enterprises – breeders of tested cattle: Black-and-White Holsteinized cattle – “Agronom”, Red Steppe cattle – “Kolos”, Ayrshire cattle – “Im. V.I. Chapaeva”. |
|||
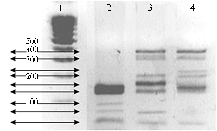
The structure of gag-gene sequences was analyzed upon the available data about 11 BLV isolates from NSBI database, and the most variable regions of gag-gene were revealed. According to these facts two endonucleases (HaeIII and FaeI) were used to assess the length polymorphism of restriction fragments. RFLP procedure was pre-optimized: PCR – in respect to the maximum yield of amplicons, RFLP – in completeness of hydrolysis, electrophoretic separation – for regime settings ensuring the highest precision of patterns. Genotyping of a limited panel of DNA from AGID-seropositive animals was performed using the samples (10 from each panel) were selected randomly, so the panel included isolates from individuals of different breeds and habitat. A sample electrophoregram of amplicon restriction products hydrolyzed by endonuclease HaeIII is shown in Figure 2.
|
Fig. 2. A sample electrophoregram of amplicon restriction products hydrolyzed by endonuclease HaeIII (347 bp) corresponding to gag-gene of BLV proviral DNA in peripheral blood leucocytes: 1 — molecular weight marker (Medigen Lab., Novosibirsk), 2-4 — experimental samples (random selection; respectively, II, X and IV conventional genotypes of the provirus). |
| The method was used to test cattle breeds bred in different farm enterprises: Black-and-White Holsteinized cattle (“Agronom”), Red Steppe cattle (“Kolos”), and Ayrshire cattle (“Im. V.I. Chapayeva”) (Krasnodar krai, 2006-2009). | |
To compare actual and expected spectra of the hydrolysis products, all studied BLV variants were separated by the polymorphism of gag-gene. Hydrolysis with restrictase HaeIII provides visualization of at least 8 groups of patterns, Fae I - 4 groups, including the variant with no restriction sites. That is, using two these restrictases allows distinguishing at least 11 conventional genotypes detected in the studied groups of cattle: 8 – in Ayrshire cattle, 6 - in Black-and-White Holsteinized and 3 – in Red Steppe cattle. In all studied animals there were recorded three identical conditional genotypes of BLV provirus - I, II and III (Table 2).
2. Distribution (%) of bovine leukemia virus conventional genotypes (for gag-gene) in surveyed farm enterprises of Krasnodar krai (2006-2009) |
|||||||||||
Enterprise, breed |
Conventional genotype |
||||||||||
I |
II |
III |
IV |
V |
VI |
VII |
VIII |
IX |
X |
XI |
|
“Agronom” – Red Steppe breed |
17 |
8 |
75 |
– |
– |
– |
– |
– |
– |
– |
– |
“Kolos” – Black-and-White Holsteinized |
44 |
12 |
6 |
6 |
13 |
19 |
– |
– |
– |
– |
– |
“Im. V.I. Chapayeva” – Ayrshire |
40 |
6 |
6 |
– |
– |
– |
7 |
20 |
7 |
7 |
7 |
On average |
36 |
10 |
27 |
2 |
5 |
7 |
2 |
7 |
2 |
2 |
2 |
Note. Dashes – the variant not identified. |
|||||||||||
Thus, the authors have studied the distribution of conditional genotypes of bovine leukemia virus (BLV) for gag-gene in three farm enterprises of Krasnodar krai. The greatest polymorphism of BLV provirus genotypes was detected in Ayrshire cattle, the lowest - in Red Steppe cattle, so it can be assumed the relationship between cattle breed and susceptibility to a particular genotype for BLV gag-gene.
REFERENCES
1. Chichinina S.V., Role of Allelic Variation of Cytokine Genes in Cattle Resistance to Bovine Leukosis, Extended Abstract of Cand. Sci. Dissertation, Novosibirsk, 2005.
2. Drobot E.V., The Study of Genotypic Variation of Bovine Leukosis Virus, Its Epizootological and Hematological Manifestation, Extended Abstract of Cand. Sci. Dissertation, Novosibirsk, 2007.
3. Ivanov O.V., Fedotov V.P., and Kryuchkova E.N., About Efficiency of Serodiagnosis of Cow Leucosis during Trematoda Invasion, S.-kh. biol., 2009, vol. 2, pp.111-113.
4. Krikun V.A., Bovine Leucosis and Immunological Tolerance, Veterinariya, 2002, vol. 6, pp. 7-9.
5. Smirnov P.N., Bolezn’ veka – leikoz krupnogo rogatogo skota (Bovine Leukemia – Cattle Disease of the Century), Novosibirsk, 2007.
6. Smirnov P.N., Gracheva N.V., and Belyavskaya V.A., Genotypic Diversification of Bovine Leukemia Virus in Different Cattle Breeds, Agrarnyi vestnik Urala, 2009, vol. 4, pp. 89-91.
7. Petropavlovsky M.V., Donnik I.M., and Tatarchyuk A.T., Metodologiya snizheniya infitsirovannosti zhivotnykh virusom leikoza v ozdoravlivaemykh stadakh krupnogo rogatogo skota (Method for Decreasing the Infestation of Cattle with Bovine Leukemia Virus during the Improvement of Herd Health), Ekaterinburg, 2009.
8. Yang D., Snanks R.D., Stewart J.A., and Lewin H.A., Milk and Fat Yields Decline in Bovine Leukemia Virus-Infected Holstein Cattle with Persistent Lymphocytosis, PNAS USA, 1993, vol. 90, pp. 6538-6541.
9. Beier D. and Siakkou H.A., Comparison of Serological Tests for the Diagnosis of Enzootic Bovine Leukosis and Eradication of Infection from a Large Herd, Tierarztliche umschau., 1994, vol. 49, no. 6, pp. 356-360.
10. Mohammadabadi M.R., Detection of Bovine Leukemia Virus Proviral DNA in Yaroslavl’, Mongolian and Black Pied Cattle by PCA, Cell. Mol. Biol. Lett., 2004, vol. 9, no. 4A, pp. 766-768.
11. Riebe R., Blankenstein P., Starick E., and Bondzio A., Establishment of a New Bovine Leucosis Virus Producing Cell Line, J. Virol. Meth., 2004, vol. 121, no. 2, pp. 239-246.
12. Johnston E.R., Albritton L., and Radke K., Envelope Proteins Containing Single Amino Acid Substitutions Support a Structural Model of the Receptor-Binding Domain of Bovine Leukemia Virus Surface Protein, J. Virol., 2002, vol. 76, no. 21, pp. 10861-10872.
13. Licursi M., Inoshima Y., Yokoyama T., Gonzales E., and Sentsui H., Genetic Heterogeneity Bovine Leukemia Virus Genotypes and Its Relation to Humoral Responses in Hosts, Virus Res., 2002, vol. 86, pp. 101-110.
Novosibirsk State Agrarian University, |
Поступила в редакцию |