УДК 636.52/58:575.174
INTRA- AND INTERBREED HETEROGENEITY AND DIVERGENCE IN FOUR HEN BREEDS, HAVING GENETIC ROOTS OF FIGHTING BIRD
A.F. Yakovlev, V.I. Tyshchenko, N.V. Dement’eva, O.V. Mitrofanova, V.P. Terletskii, A.B. Vakhrameev
By the method of DNA fingerprinting the authors determined the intra- and interbreed coefficients of genetic relation, genetic distances and the degree of heterogeneity in hens of the Yurlovskaya loud-voiced, Orlovskaya calico, Uzbekskaya fighting and Moskovskaya fighting breeds, having the genetic roots of fighting hens. The Orlovskaya calico breed is the most genetic distant from three other studied breeds. The level of heterozygosity in the hens of this breed is also the lowest. In studied breed the authors revealed the marker fragments of DNA (or their absence). The problems of genetic interrelation between four studied breeds are discussed in connection with their belonging to fighting type.
Keywords: DNA polymorphism, DNA fingerprinting, breeds of chickens, genetic distance, heterozygosity.
Molecular markers based on polymorphism of DNA and genes are becoming a key tool for genetic investigations of farm animals. A wide range of specified techniques, such as polymerase chain reaction (PCR), capillary electrophoresis and DNA fingerprinting allow studying different polymorphic DNA systems. The latter method detects polymorphism of microsatellite DNA fragments of different lengths (or molecular weight). Along with it, having the main target in routine evaluation of pedigree livestock gene funds requires considering the simplicity and cost of performed tests.
Creation of highly productive commercial breeds contributes to a gradual displacement of the initial gene pool of ancient indigenous breeds carrying valuable genetic material that often remains untapped. At the same time, these genetic resources can be demanded in future to meet possible changes of farming conditions, short-term market demands and many other factors. Unfortunately, the number of such livestock has been steadily declining and these breeds often are endangered. Preservation of rare local breeds requires maintaining a large population, which is not economically profitable. In such cases, DNA technology can greatly help to control genetic diversity in animal populations.
Polymorphic DNA markers have been used to identify chicken breeds of different origin (1, 2). The methods for assessing genetic diversity in chicken breeds and lines have been developed (3, 4) and polymorphic single nucleotide systems of the lines were analyzed in order to link them with commercial characteristics of poultry (5). Microsatellite DNA polymorphism analysis has been performed to accurately measure the degree of divergence between modern and ancient chicken breeds (6). DNA fingerprinting technology fingerprinting has been used to investigate gene funds of industrial poultry populations (7). This technique allows to evaluate genetic diversity of breeds, lines and populations, to reveal inter- and intrabreed polymorphism and to assess genetic kinship of different breeds.
This research was focused on identification of the intra- and interbreed genetic heterogeneity and diversity in chicken breeds descending from fighting chickens.
Technique. The study was performed on chickens of pedigree gene fund populations of four breeds: Yurlov Crower (Yurlov Vociferous), Russian Orloff, Uzbek Game and Moscow Game (experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). Genetic diversity of the bird was assessed by DNA fingerprinting (3) including DNA extraction, DNA enzyme restriction, electrophoresis of restriction fragments in agarose gel (“Sigma”, USA) and further transfer of them from the gel on a filter, pre-hybridization and hybridization of DNA; after detection of digoxigenin on filters, the distribution of hybridization fragments on the filters was analyzed (“Amersham”, UK) using (GTG)5 probe (5-fold repeat of GTG sequence).
The computer program Gelstats was used to calculate population-genetic characteristics – the number of bands per lane, the probability of detecting two identical genotypes (P), coefficients of genetic similarity within groups (BS1) and between groups (BS2), genetic distances (D), the number of loci, number of alleles per one locus and degree of heterozygosity (3). The sample was formed using the recommendations developed for DNA fingerprinting studies (8) providing the reliable data on a sample of 10-15 individuals.
Results. Brief description of the studied breeds suggests their origin from fighting chickens (Table 1).
1. Characteristics of the pedigree gene fund populations of chicken breeds (experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). |
|||||
Breed |
Breeds really /possibly participating the formation |
Productivity type |
Productivity characteristics |
||
Egg production, pcs/year |
Live weight, kg |
Egg weight, g |
|||
Yurlov crower |
Brahma the Malaysian fighting chickens |
Meat-egg |
160-200 |
2,6-3,0 + |
58-62 |
Russian Orloff |
Malay fighting chickens |
Ornamental - Game |
140-160 |
1,6-2,7 + |
51-52 |
Uzbek Game |
Indigenous chickens from Central Asia |
Game |
110-130 |
2,4-2,8 + |
53-57 |
Moscow Game |
Belgian and Malay fighting chickens |
Game |
150-160 |
2,2-3,0 + |
55-57 |
The obtained hybridization spectra showed high genetic diversity both within and between breeds. Interbreed polymorphism was manifested as differences in number of bands, their distribution, intensity and width of individual bands. The highest individual polymorphism was observed among DNA fragments ranging in length from 7 to 20 kbp. Both individuals and particular breeds revealed groups of similar bands on the filter area corresponding to 2-7 kbp. In general, there was observed a trend to reduce in frequency of polymorphic bands with decrease in fragment length.
The probability of detecting two birds carrying identical set of DNA fragments (P) corresponded to a single case per several billions individuals, which indicates high accuracy of the method (Table 2). Russian Orloff was found to be the breed genetically most distant from three other studied breeds.
2. Coefficients of intra- and interbreed similarity (BS) and genetic distances (D) of chickens from the pedigree gene fund populations of studied breeds descending from fighting chickens (based on data of DNA-fingerprinting; experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). |
||||||
Compared breeds |
n |
Number of bands per lane, X±m |
P |
BS1 |
BS2 |
D |
Yurlov Crower / |
10 |
37,2±2,0 |
1,8½10-18 |
0,33 |
0,28 |
0,100 |
Yurlov Crower / |
10 |
37,2±2,0 |
1,8½10-18 |
0,33 |
0,28 |
0,070 |
Yurlov Crower / |
10 |
37,2±2,0 |
1,8½10-18 |
0,33 |
0,29 |
0,050 |
Russian Orloff / |
10 |
30,1±1,1 |
5,7½10-12 |
0,42 |
0,25 |
0,140 |
Russian Orloff / |
10 |
30,1±1,1 |
5,7½10-12 |
0,42 |
0,29 |
0,090 |
Uzbek Game / |
11 |
33,4±1,5 |
1,7½10-15 |
0,36 |
0,29 |
0,060 |
Note. P – probability of presence of two individuals with identical set of DNA fragments, BS1 and BS2 — coefficients of intra- and interbreed similarity. |
||||||
Groups of Yurlov Crower and Moscow Game chicken breeds demonstrated high genetic similarity and, correspondingly, low values of genetic distance (D = 0,050). Almost similar small genetic distances were found between Uzbek Game and two breeds - Moscow Game (D = 0,060) and Yurlov Crower (D = 0,070). However, despite the significant genetic similarity between these breeds, there were differences in frequency of individual DNA fragments (Table 3).
3. The frequency of individual DNA fragments in chickens from the pedigree gene fund populations of studied breeds descending from fighting chickens (experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). |
||||
№ DNA fragments |
Yurlov Crower |
Russian Orloff |
Uzbek Game |
Moscow Game |
13 |
0,80 |
0,10 |
0 |
0,27 |
52 |
0,20 |
0,60 |
0,36 |
0,91 |
106 |
0,40 |
0,90 |
0,09 |
0,64 |
114 |
0,30 |
0,80 |
0,09 |
0,36 |
120 |
0,30 |
1 |
0 |
0,27 |
123 |
0,50 |
0 |
0,73 |
0,64 |
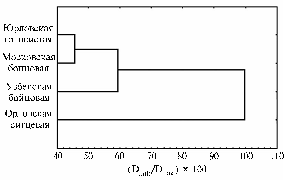 |
Dendrogram for the pedigree gene fund populations of studied chicken breeds descending from fighting chickens (based on data of DNA-fingerprinting; experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). |
Thus, despite the exchange of genetic material between Yurlov Crower and the game breeds, each of them showed distinctive features characteristic for the studied population. For example, 80% individuals of Yurlov Crower were established to carry a fragment № 13, which can be used as a marker of the breed. On the contrary, the studied population of Uzbek Game didn’t carry this fragment at all. In Russian Orloff chickens, there were specific fragments № 106 and № 120, while a fragment № 123 wasn’t detected in this breed. Genetic relationships between the studied breeds are shown on the dendrogram reflecting a somewhat isolation of Uzbek Game breed and significant isolation of Russian Orloff (Fig.).
Calculation of heterozygosity (Table 4) showed the lowest genetic heterogeneity in the studied sample of Russian Orloff chickens relative the three other studied breeds having close heterozygosity levels. It should be considered that the degree of heterozygosity reflects breeding processes in particular population.
4. Heterozygosity characteristics of chickens from the pedigree gene fund populations of studied breeds descending from fighting chickens (based on data of DNA-fingerprinting; experimental farm of the All-Russia Research and Development Institute of Farm Animal Genetics and Breeding (VNIIGRZh), Leningrad province). |
|||||||
Breed |
n |
Number |
H1 |
H2 |
H3 |
||
loci |
alleles |
polymorphic loci |
|||||
Yurlov Crower |
10 |
21,19 |
5,66 |
1,00 |
0,76 |
0,86 |
0,81 |
Russian Orloff |
10 |
18,21 |
5,11 |
0,95 |
0,65 |
0,75 |
0,71 |
Uzbek Game |
11 |
19,23 |
5,72 |
1,00 |
0,73 |
0,83 |
0,78 |
Moscow Game |
11 |
20,81 |
5,62 |
1,00 |
0,75 |
0,84 |
0,80 |
Note. Н1 — average heterozygosity (11), Н2 — corrected average heterozygosity (11), Н3 — average heterozygosity according to Jin & Chakraborty (12). |
|||||||
As mentioned above, the studied breeds were chosen for a subject of research owing to the fighting ancestors present in their pedigree. The comparative morphological study of Uzbek Game breed and 29 populations of different origins has shown the location of Russian Orloff and Uzbek Game breeds in one cluster of a cladogram (9). There’s no definite data on the origin of Russian Orloff breed although it possesses some characteristics of fighting morphological type (Table 1). The similar analysis of measurements and the frequency of alleles of biochemical loci suggests relative proximity of Uzbek and Moscow Game breeds (10). It’s hard to explain the observed similarity of Yurlov Crower and Moscow Game breeds, which though suggests the possibility of genetic crosses between these breeds in the past.
There are the prospects for development of genomic evaluation techniques providing isolation of molecular markers associated with particular characteristics and further using them to transfer polymorphic genes encoding desired traits (legs strength, type of behavior, resistance to diseases, etc.) in breeding practice (13).
Thus, the studied breeds diverged during the long-time process of breed formation, and Russian Orloff breed manifests this fact to the greatest extent. At the same time, there was revealed genetic similarity of studied breeds, which can be explained by descending these breeds from the root of fighting chickens.
REFERENCES
1. Semenova S.K., Filenko A.L. and Vasil’ev V.A., Use of Polymorphic DNA-Markers for Differentiation of Chicken Breeds of Varying Origin, Genetika, 1996, vol. 32, no. 5, pp. 795-803.
2. Tadano R., Nishibori M., Nagasaka N. and Tsudzuki M., Assessing Genetic Diversity and Population Structure for Commercial Chicken Lines Based on Forty Microsatellite Analyses, Poult. Sci., 2007, vol. 86, no. 11, pp. 2301-2308.
3. Tyschenko V.I., Dement’eva N.V., Terletskii V.P. and Yakovlev A.F., Assessment of Genetic Diversity in Poultry Populations Based of Genomic Fingerprinting, S.-kh. biol., 2002, no. 6, pp. 43-46.
4. Tyschenko V.I., Mitrofanova O.V., Dement’eva N.V. and Yakovlev A.F., Assessment of Genetic Diversity in Poultry Breeds and Experimental Populations Using DNA-Fingerprinting, S.-kh. biol., 2007, no. 4, pp. 29-33.
5. Loywick V., Bed'hom B., Pinard-van der Laan M., Pitel F., Verrier E. and Bijma P., Evolution of the Polymorphism at Molecular Markers in QTL and Non-QTL Regions in Selected Chicken Lines (Open Access Publication), Genet. Sel. Evol., 2008, vol. 40, pp. 639-661.
6. Tadano R., Nishibori M., Imamura Y., Matsuzaki M., Kinoshita K., Mizutani M., Namikawa T. and Tsudzuki M., High Genetic Divergence in Miniature Breeds of Japanese Native Chickens Compared to Red Jungle Fowl, as Revealed by Microsatellite Analysis, Anim. Genet., 2008, vol. 39, no. 1, pp. 71-78.
7. Yakovlev A., Dement’eva N., Terletskii V., Tyschenko V., Mitrofanova O.V., Tuchemskii L. and Gladkova G., Control of Genetic Diversity with the Use of DNA-Techniques, Ptitsevodstvo, 2008, no. 12, pp. 3-5.
8. Mathur P.K., Ponsuksili S. and Groen A.F., Estimation of Genetic Variability within and between Populations Using DNA Fingerprinting, Proc. of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, Canada, 1994, pp. 528-531.
9. Moiseeva I.G., Semenova S.K., Gorbacheva N.S. et al., Genetic Structure and Origin of Rare Russian Orlov Chicken Breed, Genetika, 1994, vol. 30, no. 5, pp. 681-694.
10. Moiseyeva I.G., Romanov M.N., Nikiforov A.A., Sevastyanova A.A. and Semyenova S.K., Evolutionary Relationship of Red Jangle Fowl and Chicken Breeds, Genet. Select. Evol., 2003, vol. 35, pp. 403-423.
11. Stephens J.C., Gilbert D.A., Yuhki N. and O’Brien S.J., Estimation of Heterozygosity for Single-Probe Multilocus DNA Fingerprints, Mol. Biol. Evol., 1992, vol. 9, pp. 729-743.
12. Rogstad S.H. and Pelican S., GELSTATS: a Computer Program for Population Genetics Analysis Using VNTR Multilocus Probe Data, BioTechniques, 1996, vol. 21, pp. 1128-1131.
13. Chan E.K.K., Hawken R. and Reverter A., The Combined Effect of SNP-Marker and Phenotype Attributes in Genome-Wide Association Studie, Anim. Genet., 2008, vol. 40, no. 2, pp. 149-156.
All-Russia Research and Development Institute of Farm Animas Genetics and Breeding, RAAS, St. Petersburg – Pushkin 196601, Russia, |
Received July 15, 2010
|













