УДК 636.034:575.174:575.113
POLYMORPHISM OF SOMATOTROPIN bGH AND PROLACTIN bPRL GENES AND ITS CORRELATION WITH THE MILK FAT CONTENT IN COWS OF THE KOSTROMSKAYA BREED
I.V. Lazebnaya1, O.E. Lazebnyi2, M.N. Ruzina1, G.A. Badin3, G.E. Sulimova1
In three herds of cows of the Kostromskaya breed the authors investigated the genetic structure of the somatotropin (bGH-AluI) and prolactin (bPRL-RsaI) genes with the use of PCR-RFLP (restriction fragment length polymorphism) method. The predominance of L allele of bGH gene (0.87-0.98) and A — of bPRL gene (0.67-0.87) was established in all samples. For the bGH and bPRL genes the values of observed heterozygosity (НO) are 0.05-0.07 и 0.26-0.43, respectively. Two of investigated populations differ reliably in both used DNA-markers. On the somatotropin bGH gene the VV genotype did not detected. The VL genotype, as opposed to LL genotype, reveals correlation with higher content (%) of the fat in milk.
Keywords: Kostroma cattle breed, PCR-RFLP, bPRL and bGH genes, milk production.
The prospects in using DNA markers for milk production in cattle are based on polymorphism of genes forprolactin bPRL and growth hormone bGH. These hormones regulate a large spectrum of functions in mammalians’ organism: body growth and development, start of lactation and maintaining it, as well as other effects on metabolic processes (1-3). Years of research have proven the connection between single nucleotide polymorphism (SNP) of bPRL and bGH genes and milk productivity indices - milk yield, fat and protein yield, and their percentages. Thus, it has been reported about C-G transversions in the third exon of the growth hormone gene bGH (nucleotide position 2141) followed by disappearance of restriction site for endonuclease AluI. This SNP leads to the substitution of leucine for valine at position 127 in the protein product of bGH gene known as Leu-Val polymorphism. In a number of breeds this polymorphism is correlated with milk productivity indices (4-7). Along with it, quantitative and qualitative traits of milk productivity can be affected by synonymous A-G transitions in the third exon of prolactin gene (codon 103) followed by emergence of restriction site for endonuclease RsaI (4-7). Polymorphism of these loci can be used to estimate total intra- and interbreed variability in order to correct negative effects of selection such as inbreeding (8).
Kostroma breed is one of three breeds of brown cattle bred in Russia. Its features are intense growth, strong constitution, stable inheritance of characteristics, high quality milk. Kostroma cattle is considered as one of the best productive breeds used for multiple purposes. Traditionally, this breed is bred in Kostroma, Vladimir and Ivanovo provinces of the Russian Federation and in the Republic of Belarus. In recent decades, there was observed an unfortunate trend to reduce in its number: from 1980 to 2004, population of Kostroma cattle decreased by 4,3 times and amounted to 193,2 thousand animals. (9).
Genetic structure of the Kostroma breed was investigated using different types of markers, including markers for blood proteins, for genes of milk proteins - casein and lactalbumin, etc. (10-14). However, there’s almost no data on polymorphism of genes bPRL and bGH in this breed (14).
The purpose of this research was selected considering the decrease in number of Kostroma livestock and the insufficient knowledge about its genetic potential for key selection markers: to evaluate the intrabreed variability in Kostroma cattle using polymorphism of genes for growth hormone bGH (AluI) and prolactin bPRL (RsaI), and to reveal its correlation with milk fat content.
Technique. The polymorphism of genes bPRL and bGH was analyzed using PCR-RFLP in cows of Kostroma breed bred in pedigree farms of several enterprises: the Experimental Production Enterprise “Minskoe” (Kostroma municipal district) (n = 20, 2008), the Agricultural Producers’ Co-operative ‘Gridino” (Krasnoselsky district of Kostroma province) (n = 42, 2008) and the Regional State Unitary Enterprise the Pedigree Farm “Luzhki” (Nerekhtsky district of Kostroma province) (n = 62, 2006). DNA was extracted from the whole blood (200 ml, a set of reagents DIAtomTM DNA Prep by “Isogene Lab. Ltd.”, Russia). Amplification of gene fragments bPRL (156 bp) and bGH (223 bp) was performed according to standard techniques, as described previously (15), using a set GenePakTM PCR Core (“Isogene Lab. Ltd.”, Russia) in the thermocycler Tertsik (“DNA-Technology”, Russia). Restriction endonucleases RsaI and AluI were applied following the manufacturer’s recommendations (“MBI Fermentas”, Lithuania).
Statistical analysis was performed using the programs Popgene v.1.31 and Statistica 6.0. (16, 17).
Results. BB genotype for prolactin gene bPRL was not found in "Minskoe"; in “Luzhki” it occurred with very low frequency equal to 3,2% and in “Gridino” it was detected about 4 times more frequently.
1. The frequency of genotypes and alleles of genes for prolactin bPRL and growth hormone bGHin studied populations of Kostroma cattle bred in pedigree farms of different enterprises (Kostroma province.) |
||||
Enterprise |
Genotype on a gene |
Genotype frequency ±se |
Allele |
Allele frequency±se |
Gene bPRL |
||||
“Luzhki” |
AA |
0,581±0,063 |
A |
0,774±0,043 |
AB |
0,387±0,062 |
B |
0,226±0,079 |
|
BB |
0,032±0,022 |
|
|
|
“Gridino” |
AA |
0,452±0,077 |
A |
0,667±0,063 |
AB |
0,429±0,076 |
B |
0,333±0,089 |
|
BB |
0,119±0,050 |
|
|
|
“Minskoe |
AA |
0,737±0,101 |
A |
0,868±0,059 |
AB |
0,263±0,101 |
B |
0,132±0,151 |
|
BB |
0 |
|
|
|
Gene bGH |
||||
“Luzhki” |
VV |
0 |
V |
0,081±0,086 |
VL |
0,161±0,047 |
L |
0,919±0,026 |
|
LL |
0,839±0,047 |
|
|
|
“Gridino” |
VV |
0 |
V |
0,131±0,102 |
VL |
0,262±0,068 |
L |
0,869±0,039 |
|
LL |
0,738±0,068 |
|
|
|
“Minskoe |
VV |
0 |
V |
0,025±0,156 |
VL |
0,050±0,049 |
L |
0,975±0,025 |
|
LL |
0,950±0,049 |
|
|
|
Note. se — standard error of data for frequencies of a genotype and allele |
||||
The studied samples showed different ratios of AA and AB genotypes as well (Table 1). Thus, in “Gridino” they were present with equal frequency, while in two other enterprises AA genotype was dominant: in “Minskoe”, its frequency reached about 74%, in “Luzhki” - 58%. Such ratio of genotypes influenced the distribution of allele frequencies: A allele was most frequent in all samples and its rate in “Minskoe”, “Luzhki” and “Gridino” amounted to, respectively, 87, 77 and 67%.
In the studied groups of cattle, there were revealed no carriers of VV genotype for the gene bGH, the low percentage of heterozygotes and the high frequency of LL homozygotes. Most contrast ratio of genotypes VL and LL was found in “Minskoe” (respectively, 5 and 95%), less contrast – in “Luzhki” (16 and 84%) and in “Gridino” (26 and 74%). Distribution of alleles with high frequency of allele L was similar in all the farms (Table 1).
The established frequencies of genotypes were verified with the Hardy-Weinberg principle and all the analyzed samples for each gene were found to be present in an equilibrium. For bGH gene in “Minskoe” G = 0, d.f. = 1, P = 1,000; in “Gridino” G = 1,513, d.f. = 1, P = 0,219; in “Luzhki” G = 0,791, df = 1, P = 0,374. For bPRL gene in “Minskoe” G = 0,888, d.f. = 1, P = 0,346; in “Gridino” G = 0,096, d.f. = 1, P = 0,757; in “Luzhki” G = 0,664, d.f. = 1, P = 0,415.
2. The observed (HO) and expected (HE) heterozygosity for prolactin gene bPRL and growth hormone gene bGH in studied groups of Kostroma cattle bred in pedigree farms of different enterprises (Kostroma province.) |
|||
Enterprise |
Gene |
HO |
HE |
“Luzhki” (n = 62, 2006) |
bPRL |
0,387±0,060 |
0,353±0,161 |
“Gridino”(n = 42, 2008) |
bPRL |
0,429±0,076 |
0,444±0,073 |
“Minskoe”(n = 20, 2008) |
bPRL |
0,263±0,101 |
0,229±0,170 |
“Luzhki” (n = 62, 2006) |
bGH |
0,161±0,047 |
0,150±0,152 |
“Gridino”(n = 42, 2008) |
bGH |
0,262±0,068 |
0,228±0,173 |
“Minskoe”(n = 20, 2008) |
bGH |
0,050±0,049 |
0,049±0,060 |
In the studied groups of cattle, the values of observed (HO) and expected (HE) heterozygosity (Table 2) for each of the markers didn’t differ reliably. In general, HO for prolactin gene bPRL was higher than for growth hormone gene bGH. These indices for both markers were found to decrease in the line: ”Gridino” – “Luzhki” – “Minskoe”.
Results of G-test (probability) (Table 3) indicate significant differences on the both markers between groups of cattle from "Minskoe” and “Gridino”, while none of them differed from “Luzhki”.
3. Reliability of differences in the distribution of genotype frequencies for prolactin gene bPRL and growth hormone gene bGH in studied groups of Kostroma cattle bred in pedigree farms of different enterprises (Kostroma province.) |
|||
Enterprise |
“Gridino |
“Minskoe” |
“Luzhki” |
“Gridino”(n = 42, 2008) |
|
0,047 |
0,174 |
“Minskoe”(n = 20, 2008) |
0,035 |
|
0,387 |
“Luzhki” (n = 62, 2006) |
0,219 |
0,180 |
|
Note. Probability values (G-test) of the genotypes for genes bPRL and bGH are located respectively above and below the diagonal. |
|||
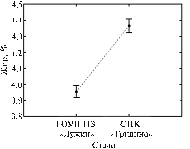
|
The average fat content in milk from Kostroma cows bred in two enterprises (vertical lines – 95% CI) (Kostroma province, 2006, 2008). |
In the studied groups of Kostroma cattle, interpopulation variability (FST = 0,026) for bGH gene was significantly lower than intrapopulation variability (0,050; 0,161 and 0,262 in “Gridino”, “Luzhki” and “Minskoe”, resp.). FST for bPRL gene amounted to 0,031, which was also considerably lower than intrapopulation variability in studied groups (0,263; 0,387 and 0,429 in “Gridino”, “Luzhki” and “Minskoe”, resp ").
The multivariate dispersion analysis of a total sample revealed the impossibility of simultaneous considering four factors (genotypes for genes bGH and bPRL, belonging to the line and to the herd). Upon this fact, there were excluded the lines represented by an only herd or by an only animal; thus, the herd from “Minskoe” wasn’t analyzed as it included only one line common with two other enterprises. The three-factor analysis of variance considering the influence of each separate factor (gene, line and enterprise) has given the following results: for bPRL gene, the gene factor - P = 0,816, the line factor - P = 0,364, and the herd factor - P = 0,001; for bGH gene , the gene factor - P = 0,086, the line factor - P = 0,236, and the herd factor - P = 0,001.
These probabilities indicate that fat content in milk from Kostroma cows in correlated with the only factor - belong to the herd (Fig.). Higher values of fat content in milk were observed in animals from the herd of “Gridino” (Fig.).
Despite the absence of reliable correlation between the genotype for the studied genes and fat content in milk, a comparison of probabilities of both genes’ influence on this trait suggests a small trend to the correlation between bGH gene and fat content in milk. At the same time, there are the literature data about specific effects of certain genotypes on fat content in milk from cows of different breeds. According to H. Yardibi et al. (18), in Eastern Anatolian Red and the South Anatolian Red cows, LL genotype (in contrast to genotypes VL and VV) is highly correlated with a higher fat content in milk. The data on effects of bPRL gene show that a number of cattle breeds demonstrate fat content of milk influenced by genotypes detected by RsaI restriction endonuclease (in contrast to Kostroma cattle). For example, Holstein-Friesian cows carrying AA genotype produce milk with higher fat content than those with BB genotype(6). On the contrary, Russian Black-and-White and Red-and-White breeds demonstrate the highest fat content in milk from cows with BB genotype and heterozygotes (19, 20).
Have analyzed bPRL(RsaI)- and bGH(AluI)- variability in the studied groups of Kostroma cattle, the authors revealed the distribution of alleles for prolactin gene similar to that in a number of European breeds (8). For example, the frequency of A-allele in Czech, Polish and German Red breeds are respectively 0,559; 0,869 and 0,909. The frequency of V-allele of growth hormone gene of was very low in Kostroma cows from “Minskoe” (0,025), and somewhat higher – in “Luzhki” (0,080), which corresponds to the values ??of Danish Jersey breed (0,070 ) (21). Only in "Gridino" the frequency of V-allele (0,130) was comparable to that in German Red breed (0,150) (8), but lower than in Lithuanian Black-and-White and Red breeds (0,300 and 0,230, resp.) (22). The outstandingly high frequency of V-allele was observed in Czech Red cattle (0,510) (8).
Thus, three groups of Kostroma cattle were analyzed using DNA markers and two of them bred in the farms of “Gridino” and “Minskoe” were found to have reliably different frequencies of alleles and genotypes for both bPRL and bGH genes. There was established a low interbreed diversity assessed by heterozygosity for bGH gene, and the average degree – for bPRL gene compared to that in a number of domestic breeds (15, 23). The diversity for both markers was found to decrease in the line: “Gridino” – “Luzhki” – “Minskoe”. In the studied Kostroma cows, the analysis of correlation between fat content in milk and polymorphism of bPRL and bGH genes considering the factors of herd and line (on father) revealed the reliable effect only from belonging to the herd. At the same time, many Russian and foreign authors have reported about the correlation of certain genotypes for these genes with productivity indices in a number of breeds and it was suggested using these data in breeding practice. Findings of this research has shown that implementation of such approach requires a more detailed investigation of the gene pool of domestic breeds.
REFERENCES
1. Burton J.L., McBride B.W., Block E. et al., A Review of Bovine Growth Hormone, Can. J. Anim. Sci., 1994, vol. 74, pp. 167-201.
2. Ohlsson C., Bengtsson B., Isaksson O. et al., Growth Hormone and Bone, Endocr. Rev., 1998, vol. 19, pp. 55-79.
3. Leprovost F., Leroux C., Martin P. et al., Prolactin Gene Expression in Ovine and Caprine Mammary Gland, Neuroendocrinology, 1994, vol. 60, pp. 305-313.
4. Chrenek P., Huba J., Oravcova M. et al., Genotypes of Bgh and Bprl Genes in Relationships to Milk Production, Proc. EAAP 50th Annual Meeting (Book of Abstracts), Zurich, 1999, p. 40.
5. Mitra A., Schlee P., Balakrishnan C.R. et al., Polymorphisms at Growth Hormone and Prolactin Loci in Indian Cattle and Buffalo, J. Anim. Breed. Genet., 1995, vol. 112, pp. 71-74.
6. Chung E.R., Rhim T.J. and Han S.K., Associations between PCR-RFLP Markers of Growth Hormone and Prolactin Genes and Production Traits in Dairy Cattle, Korean J. Anim. Sci., 1996, vol. 38, pp. 321-336.
7. Dybus A., Grzesiak W., Szatkowska I. et al, Association between the Growth Hormone Combined Genotypes and Dairy Traits in Polish Black-and-White Cows, Anim. Sci. Pap. Rep., 2004, vol. 22, no. 2, pp. 185-194.
8. Zato-Dobrowolska M., Citek J., Filistowicz A. et al., Genetic Distance between the Polish Red, Czech Red and German Red Cattle Estimated Based on Selected Loci of Protein Coding Genes and DNA Microsatellite Sequences, Anim. Sci. Pap. Rep., 2007, vol. 25, no. 1, pp. 45-54.
9. Sidorenko V.A. and Baranova N.S., Kostromichi i “kostromichka”: k 60-letiyu sozdaniya kostromskoi porody krupnogo rogatogo scota (1944-2004) (Kostromichi and “Kostromichka”: a Book Dedicated to the 60-Year Anniversary of Kostroma Cattle (1944-2004), Kostroma, 2004.
10. Emel’yanov E.G., Gene Fund for the Three Types of Milk Proteins in Population of Kostroma Cattle, Nauch. tr. Leningradskogo SKhI, 1979, vol. 379, pp. 70-74.
11. Yancheva R.S., Blood Groups, Types of Hemoglobin and Transferrin in Kostroma Cattle and Opportunities for Its Use in Breeding, Extended Abstract of Cand. Sci. Dissertation, Dubrovitsy, 1970.
12. Baranov A.V., Genetic Labeling and Its Use in Improvement of Dairy Cattle Breeding, Extended Abstract of Doct. Sci. Dissertation, Moscow, 1997.
13. Glushenko M.A., Using Genetic Methods for Evaluation of Industrial Populations of Kostroma Cattle, Extended Abstract of Cand. Sci. Dissertation, Lesnye Polyany, 1999.
14. Sulimova G.E., Lazebnaya I.V., Shtyfurko T.A., Ryabinina O.M., Ukhanov S.V. and Badin G.A., Analysis of the Gene Fund of Kostroma Cattle Breed Using DNA-Markers of Genes for Kappa-Casein, Prolactin, Growth Hormone and BoLA-DRB3, in Mat. mezhvuz. nauch.-pract. konf. “Aktual’nye problemy nauki v APK” (Proc. of Higher Schools’ Sci.-Pract. Congress “Urgent Problems of Science in Agriculture”), Kostroma, 2008, vol. 3, pp. 183-185.
15. Lazebnaya I.V., Lazebnyi O.E. and Sulimova G.E., Studying Genetic Variation in Yakutian Cattle (Bos taurus L.) Using the Prolactin bPRL, Growth Hormone bGH and Transcription Factor bPit-1 Genes, Genetika, 2010, vol. 46, no. 3, pp. 425-428.
16. Yeh F.C. and Boyle T.J.B., Population Genetic Analysis of Co-Dominant and Dominant Markers and Quantitative Traits, Belgian J. Bot., 1997, vol. 129, p. 157.
17. StatSoft, Inc. STATISTICA for Windows (Computer Program Manual). Tulsa, OK: StatSoft, Inc., 1999. WEB: http://www.statsoft.com.
18. Yardibi H., Hosturk G.T., Paya I. et al., Associations of Growth Hormone Gene Polymorphisms with Milk Production Traits in South Anatolian and East Anatolian Red Cattle, J. Anim. Vet., 2009, vol. 8, no. 5, pp. 1040-1044.
19. Alipanah M., Kalashnikova L. and Rodionov G., Association of Prolactin Gene Variants with Milk Production Traits in Russian Red Pied Cattle, Iranian J. Biotechnol., 2007, vol. 5, no. 3, pp. 158-161.
20. Goryacheva T.S. and Goncharenko G.M., Polymorphism in Kappa-Casein and Prolactin Genes and Their Influence on Dairy Productivity of Cows of the Black-and-White Breed, S.-kh. biol., 2010, no. 4, pp. 51-54.
21. Sørensen P., Grochowska R., Holm L. et al., Polymorphism in the Bovine Growth Hormone Gene Affects Endocrine Release in Dairy Calves, J. Dairy Sci., 2002, vol. 85, pp. 1887-1893.
22. Skinkytè R., Zwierzchowski L., Riaubaitè L. et al., Distribution of Allele Frequencies Important to Milk Production Traits in Lithuanian Black-and-White and Lithuanian Red Cattle, Veterinarija ir Zootechnika, 2005, vol. 31, no. 53, pp. 93-97.
23. Lazebnaya I.V., Lazebny O.E., Khatami S. et al., Investigation of bPRL, bPitI, and bGH Polymorphisms in Four Native Russian Cattle Breeds, in Proc. XX International Congress of Genetics “Genetics: Understanding Living Systems”, Berlin, German Genetics Society, 2008, p. 82.
1N.I. Vavilov Institute of General Genetics, RAS, Moscow 119991, Russia, |
Received September 6, 2010
|