ÓÄÊ 635.649:631.523:57.088.1
MOLECULAR LABELING OF THE LINES AND OBTAINED ON THEIR BASIS THE VARIETIES OF Capsicum annuum L.
N.N. Ryzhova1, E.Z. Kochieva1, O.N. Pyshnaya2
On the variants of Capsicum annum L. (the L-Zolotoe Chudo, L-Beke, L-Artek lines and obtained of their basis the Snegir’ and Yantar’ varieties) with the use of appropriate primers the authors for the first time investigated the multipoint RAPD method in respect to the analysis of inheritance of parent genetic material during the varieties obtaining and establishing of their genetic nature. As result of DNA-spectra comparison the genetic polymorphism was estimated and the DNA-markers potential suitable for genetic typing of investigated variants of C. annuum were identified, the recombinant nature of the Snegir’ variety was shown and the high similarity of the Yantar’ variety with L-Artek parent line was confirmed.
Key words: pepper cultivars, hybrids, molecular DNA-markers, multiloci analysis, genetic diversity, pepper breeding.
Success in breeding is connected to many determinant factors: the proper choice of parental components, the choice of hybridization and selection methods, and, to a large degree, the parental forms’ ability to provide the combination of anticipated determinants, i.e., a genetic nature of source material (lines and hybrids obtained upon these lines) (1-2). A population formed by the planned hybridization of varieties and lines, is the most genetically valuable material for creation of new varieties and parental forms. The selection in a hybrid population starts from the identification of homozygous and heterozygous plants manifested in F2 as a result of F1 gametes fertilization (3).
The assessment of hybrid population structure and identification of potentially valuable genotypes can be performed using the modern highly accurate and reliable DNA-labeling molecular systems (so-called marker assisted selection, MAS), which significantly accelerate and simplify the procedure of selection the anticipated genotype and its subsequent tests in a selective background (4).
The development and practical use of DNA-labeling is particularly relevant in regard to the selection of vegetable crops. The protein markers effective in selection of cereals are unreliable for most vegetables due to the specific regulation of reserve proteins expression, which is highly dependent on external factors (4-5 ).
The development of DNA-markers system can be performed by modern methods of genotype molecular identification based on PCR-technology: AFLP (amplified fragment length polymorphism), DNA-RAPD (random amplification of polymorphic DNA), ISSR (inter simple sequence repeats), SSAP (sequence-specific amplification polymorphisms), SSR (simple sequence repeats), SNP (single nucleotide polymorphism) and several other methods. RAPD is one of the simplest and accessible methods allowing exploring mainly the selectively neutral unique and moderately repetitive sequences of plant genome (6). This method is widely used for varieties identification, evaluation of intervarietal polymorphism, varietal purity and varietal biotypic composition, as well as for analysis of hybrid populations, for hybridization monitoring and genetic material introgression in intervarietal, interlinear and interspecific crosses (5, 7-15).
The purpose of this work was studying the possibility of applying the multipoint RAPD method for establishment the genetic nature of sweet pepper varieties and for analyzing the inheritance of parental genetic material in newly created varieties. The following tasks were determined: selection of primers detecting polymorphism in parental lines and obtained varieties, labeling of parental lines and obtained varieties, evaluation of molecular and genetic polymorphism of studied samples, identification of DNA-markers potentially suitable for genotyping of the studied and other lines and varieties of Capsicum annum L.
Methods. The objects of the investigation were three parental lines of sweet pepper C. annuum: the L-Zolotoe Chudo, L-Beke, L-Artek lines and the Snegir’ and Yantar’ varieties obtained by simple crosses of the lines (L-Zolotoe Chudo x L-L-Beke and L-Zolotoe Chudo õ L-Artek, resp.) with a consequent selection from a hybrid population by pedigree method.
DNA was isolated from 8-10-day-old seedlings by a standard method (16) with additional deproteinization by phenol - chloroform mixture. For PCR-test, 20 primers the series OPA, OPD, OPH, OPN (“Operon Technologies”, USA) were used. Amplification was performed in a reaction mixture the volume of 15 ml containing 2,5 mM MgCl2, and 0,2 mM of each dNTP, 0,5 mM primer, 0,3 units Taq-polymerase, 1 õ buffer from the corresponding set (“Dialat Ltd.”, Russia) and 100 ng genomic DNA. The thermocycler (amplifier) GeneAmp PCR System2700 (“Applied Biosystems”, USA) was operated under the regime: denaturation - 30 s at 94 °C, primer annealing – 45 s at 37 °C (number of cycles - 36); DNA synthesis - 1 min at 72 °C, preliminary denaturation - 5 min at 94 °C, final elongation – 10 min at 72 °C.
All the reactions were performed in two replications. Amplification products were separated by electrophoresis in 1,7% agarose gel (high resolution, “Sigma”, MetaPhor, “Cambrex”, USA) in 1 x TVE-buffer followed by staining with ethidium bromide.
The clear, reproducible fragments ranging in size from 300 to 2000 bp were accounted. The polymorphism of genome amplified fragments was estimated as the ratio: (number of polymorphic fragments) / (total number of derived fragments), expressed as a percentage.
Results. To identify genetic differences between parental forms, 20 primers the series OPA, OPD, OPH, OPN, were tested, and only two of them (OPE1 and OPE7) provided clear reproducible DNA-spectra of amplification products containing specific fragments. A total of 34 RAPD-DNA fragments were obtained, 16 (47%) of which were polymorphic. The length of amplified RAPD-fragments varied within 340-1650 bp. At the same time, for the parental pair L-Beke - L-Zolotoe Chudo, the fragments OPE11550 (A), OPE11280 (B), OPE1530 (E), OPE1385 (H), OPE71150 (A´), OPE7800 (B´), OPE7720 (C´), OPE7410 ( G´) have proved to be the specific markers for L-Beke line (Fig.), and the fragments OPE1600 (C), OPE1550 (D), OPE1490 (F), OPE1410 (G), OPE1380 (I), OPE7630 (D´), OPE7410 (G´) – the markers for L-Zolotoe Chudo. Respectively, for the parental pair L-Zolotoe Chudo - L-Artek, the fragments OPE1600 (C), OPE1490 (F), OPE1410 (G), OPE1380 (I), OPE7630 (D´) appeared to be the specific markers for L-Zolotoe Chudo. For the L-Artek line, no specific amplicons were identified.
Both these primers (OPE1 and OPE7) were used for subsequent labeling of the Snegir’ an Yantar’ varieties obtained by interline crosses, for identification their genetic nature and comparative multipoint RAPD-analysis of all the five samples of pepper.
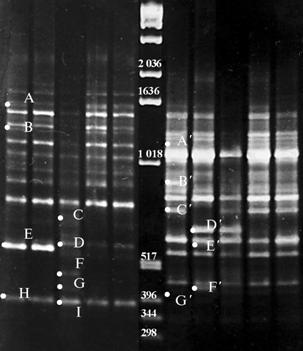 |
DNA RAPD-spectra of the parental lines of sweet pepper Capsicum annum L.. L-Beke, L-Zolotoe Chudo, L-Artec (1, 2, and 3, resp.), and the varieties Snegir’ and Yantar’ created upon these lines (4 and 5, resp.) obtained using the primers OPE1 and OPE7 (respectively, to the left and to the right from the molecular mass marker) (M, 1 kb Ladder, “Gibco BRL”, USA). |
Analyzing DNA of the L-Beke, L-Zolotoe Chudo lines and the Snegir’ variety created upon these lines, both primers (OPE1 and OPE7) were used to detect differences among the three samples. In the variant with the OPE1 primer, RAPD-spectra of the Snegir’ variety was more similar to the spectra of the L-Beke line: the fragments OPE1530 (E), OPE1385 (H) specific for the L-Beke line were observed and no one specific for L-Zolotoe Chudo. On the contrary, when testing OPE7, RAPD-spectra of the Snegir’ variety was similar to the one of the L-Zolotoe Chudo line and included its marker fragment OPE7630 (D´), while the marker fragments of the L-Beke line OPE71150 (A´) OPE7800 (B´), OPE7720 (C´), OPE7410 (G´) were absent. In RAPD-spectrum of the Snegir’ variety, the presence of own specific fragments - OPE7580 (E´) and OPE7430 (F´) was found, and the absence of some spectra fragments detected in parental lines (see Fig.), probably, as a result of recombination.
It should be noted, that Snegir’ and Yantar’ were obtained by individual selection of plants in splitting generations with pedigree accounting of the selected plants till the formation of homozygous lines. In progeny of each plant, the splitting by determinants leads to the greatest number of determinants combinations in F2 generation as the result of genetic recombination. In F2 generation, the genotypes keeping combinations of parental determinants (as was estimated by phenotype) were selected with a particular attention to economically valuable properties. In F3, the plants with anticipated recombinations of determinants were selected, especially in offspring best suited for breeding purposes, because F3 is a start level for lines formation. When creating the Snegir’ variety, the selection from hybrid population was performed by phenotype upon the determinants manifested in hybrid plants only, and some desirable properties of parental forms as well. All these data confirm our assumption about the recombinant nature of Snegir’.
The size of polymorphic RAPD-fragments in spectra obtained with the primers OPE1 (A-I) and OPE7 (A´-G´) in the studied samples of sweet pepper Capsicum annum L. |
|||||||||
Line, variety |
A/A´ |
B/B´ |
C/C´ |
D/D´ |
E/E´ |
F/F´ |
G/G´ |
H |
I |
Primer OPE1 |
|||||||||
L-Beke |
1550 |
1280 |
|
|
530 |
|
|
385 |
|
Snegir’ |
|
|
|
|
530 |
|
|
385 |
|
L-Zolotoe Chudo |
|
|
600 |
550 |
|
490 |
410 |
|
380 |
Yantar |
1550 |
1280 |
|
550 |
|
|
|
385 |
|
L-Artek |
1550 |
1280 |
|
550 |
|
|
|
385 |
|
Primer OPE7 |
|||||||||
L-Beke |
1150 |
800 |
720 |
|
|
|
410 |
|
|
Snegir’ |
|
|
|
630 |
580 |
430 |
|
|
|
L-Zolotoe Chudo |
|
|
|
630 |
|
|
410 |
|
|
Yantar |
1150 |
|
720 |
|
|
|
410 |
|
|
L-Artek |
1150 |
|
720 |
|
|
|
410 |
|
|
The RAPD-spectra comparative analysis of the L-Zolotoe Chudo, L-Artek parental forms and the Yantar’ variety has shown the similarity of the Yantar’ variety with its father component (the L-Artek line) and no fragments specific for its another parent – the L-Zolotoe Chudo line (OPE1600 - C, OPE1490 - F, OPE1410 - G, OPE1380 - I, OPE7630 - D´, OPE7410 - G´). A phenotype (habit, leaf shape and size, fruit shape, fruit position, etc.) of Yantar’ clearly repeats the one of L-Artek except the properties of precocity and fruit color, inherited from the maternal line.
Thus, applying RAPD-method, the molecular labeling of five samples of sweet pepper Capsicum annum L. (three lines and two varieties created upon the lines) was performed. RAPD-primers for detection of intervarietal polymorphism were isolated, the specific markers of RAPD-fragments were identified and the possibility of using RAPD-markers for the analysis of hybrid combinations and detection of recombination processes was shown.
REFERENCES
1. M a m e d o v M.I., P y sh n a ya O.N., D z h o s E.A. Selektsiya pertsa sladkogo dlya otkrytogo grunta Nechernozem'ya. Kartofel' i ovoshchi, 2005, 4: 14.
2. M a m e d o v M.I., P i v o v a r o v V.F. Selektsiya tomata, pertsa i baklazhana na adaptivnost'. M., 2002.
3. B u n i n M.S., M a m e d o v M.I., S h m y k o v a N.A.,
S u p r u n o v a T.P., E n
g a l y ch e v a I.A., K o c h i e v a E.Z.,
R y z h o v a N.N., D z h o s E.A.. Mezhvidovaya gibridizatsiya v rode Capsicum L. i ee ispol'zovanie v selektsii. Metodika. M., 2008.
4. L e f e b v r e V. Molecular markers for genetics and breeding: development and use in pepper (Capscum spp.). In: Molecular marker systems in plant breeding and crop improvement (N. Lorz, G. Wenzel, eds.) — Biotechnology in Agriculture and Forestry, 2004, 55: 189-214.
5. K o c h i e v a E.Z. Ispol'zovanie metodov na osnove polimeraznoi tsepnoi reaktsii dlya markirovaniya genoma rastenii. S.-kh. biol., 1999, 1: 1-19.
6. W i l l i a m s J.G.K., K u b e l i k A.R., L i v a k K.J., R a f a l s k i J.A., T i n g e y S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl. Acids Res., 1990, 18: 6531-6535.
7. C r o c k e t t P.A., B h a l l a P.L., L e e C.K., S i n g h M.B. RAPD analysis of seed purity in a commercial hybrid cabbage (Brassica oleracea var. capitata) cultivar. Genome, 2000, 43: 317-321.
8. I l b i H. RAPD markers assisted varietal identification and genetic purity test in pepper, Capsicum annuum. Scientia Horticulturae, 2003, 97(4): 211-218.
9. S u n G., W a n g - P r u s k i G., M a y i c h M., J o n g H. RAPD and pedigree-based genetic diversity estimates in cultivated diploid potato hybrids. Theor. Appl. Genet., 2003, 107: 110-115.
10. K a r i h a l o o J.L., D w i v e d i Y.K, G a i k w a d A.B. Analysis of genetic diversity of Indian mango cultivars using RAPD markers. J. Hortic. Sci. Biotechnol, 2003, 78(3): 285-289.
11. S i n g h N., S i n g h M., K u m a r S. e.a. RAPD markers for hybrid seed purity testing in tomato (Solanum lycopersicum L.). Current Science, 2007, 93(4): 462-463.
12. C h u n d e t R., C u t l e r R.W., T a s a n o n M.S. e.a. Hybrid detection in Lychee (Litchee chinensis Sonn.) cultivars using HAT-RAPD markers. ScienceAsia, 2007, 33: 307-311.
13. Î n t o S., L a o s a t N., S u k s a w a t W., P o p l u e c h a i S.,
E u n g w a n i c h a
y a p a n t P.D., C h u k e a t i r o t e E. Phylogenetic analysis of Cucumis sativus using RAPD molecular markers. J. Plant Sci., 2008, 3(1): 105-110.
14. E r c i s l i S., O r h a n E., H i z a r c i Y., Y i l d i r i m N., A g a r G. Genetic diversity in grapevine germplasm resources in the Coruh Valley revealed by RAPD markers. Biochem. Genet., 2008, 46: 590-597.
15. T h a w a r o S., T e - Ñ h a t o S. RAPD (random amplified polymorphic DNA) marker as a tool for hybrid oil palm verification from half mature zygotic embryo culture. J. Agricult. Technol., 2008, 4(2): 165-176
16. E d w a r d s K., J o h n s t o n e C., T h o m p s o n C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucl. Acids Res., 1991, 19(6): 1349.
1Center “Bioengineering”, Russian Academy of Sciences, |
Received November 25, 2009
|













