УДК 636.1:575.17:577.29
POLYMORPHISM OF MICROSATELLITE DNA IN HORSES OF STUD AND LOCAL BREEDS
V.V. Kalashnikov, L.A. Khrabrova, A.M. Zaitsev, M.A. Zaitseva, L.V. Kalinkova
Molecular-genetic peculiarities were studied in horses of the fifteen stud and local breeds multiplied in our country. The animals were evaluated using 17 microsatellite DNA loci. The private alleles were revealed in allelofond of majority breeds. The genetic distances were calculated between populations. The significant genetic differences were showed between local and stud horse breeds.
Keywords: genetic diversity, horse breeds, microsatellite DNA.
Using molecular genetic markers for studying populations and species opens up new possibilities for solving many theoretical and practical issues related to evolution of species, formation of breeds and assessment of genetic resemblance of animals at individual and population levels. The efficiency of microsatellite markers was proved by studies performed since the mid-1990s (1-2). High variability, codominant inheritance and known location in the genome make them the best tool for studying genetic characteristics and origin of species (3-6).
According to the international Food and Agriculture Organization (FAO), Russia possesses one-tenth of horse pedigree resources in the world. State register of breeding achievements includes 40 horse breeds, half of which are unique native breeds such as Orlov trotter, Russian Don, Buddyonny, Terskaya. Local breeds are interesting for researchers from different countries as well - Altai, Bashkir, Buryat, Vyatka, Transbaikalian, Kalmykian, Mezen, Pechora, Tuvan, Yakut and other populations adapted to survival in adverse climate of the Far North, Kalmykia and Siberia.
It’s important to study and maintain native horse breeds because of their good adaptation to local climatic conditions and resistance to diseases. As a rule, these breeds have a unique gene pool, which was confirmed by data on their genetic structure (7-9). Certain domestic breeds are small in number and have a critical status, so using genetic methods becomes increasingly important for preservation of such species (10).
The purpose of this research was the comparative analysis of molecular-genetic characteristics of farm and local horse breeds bred in Russia.
Technique. Polymorphism of microsatellite DNA was evaluated in 1169 horses of 15 breeds. The group of farm breeds included Arabian horse (n = 319), Thoroughbred riding (n = 443), Akhal-Teke (n = 109), Trakehner (n = 27), Orel trotter (n = 99), Russian (n = 14), Standardbred ( n = 50), French trotter (n = 13); a group of local breeds - Altai (n = 29), Buryat (n = 11), Vyatka (n = 10), Transbaikalian (Zabaikal’skaya) (n = 11), Mezen (n = 12 ), Tuvan (n = 11) and Khakassian (n = 11). The animals were tested for 17 microsatellite DNA loci included in the primer set StockMarks® (“Applied Biosystems”, USA).
DNA was extracted from blood samples and hair follicles using a kit of DiatomTM DNA Prep 200 ExtraGeneTM DNA Prep 200 (“Isogene Laboratory”, Russia). The samples were amplified in the thermocycler 2720 Thermal Cycler (“Applied Biosystems”, USA) according to manufacturer's recommendations. Electrophoresis of amplification products was carried out in the automatic 4-capillar genetic analyzer ABI 3130 (“Applied Biosystems”, USA). Genotyping of horses and graphic interpretation of genotyping profiles were carried out using a control sample of the primer kit StockMarks and results of participation in the international Horse Comparison Test (Edinburgh, 2010).
Genetic and population analysis included the assessment of polymorphism degree (Ae), observed (Ho) and expected (He) heterozygosity, counting the frequency of alleles of microsatellite loci, fixation index (Fis) and cluster analysis of populations using Microsoft Excel 2003 and Statistica 6.0.
Results. Testing the horses for each of the 17 microsatellite loci allowed identifying from 6 to 15 alleles. Three loci were found to include the alleles not described before: AHT4F – only in Akhal-Teke, ASB17W - in Altai, ASB23G and ASB23H - in Tuvan horses. The widest range of alleles of microsatellite loci (140 alleles for 17 loci) and the maximum number of private alleles (15 alleles) were detected in Akhal-Teke – one of the oldest horse breeds in the world. The minimum number of alleles were recorded in local Altai horses: 129 alleles, three of which are located in two microsatellite loci (ASB17W,V and AHT5P) and not found in other breeds (Tables 1 and 2).
1. The genetic spectrum of alleles of microsatellite loci in farm horse breeds bred (n = 1169) |
||||||||||||||
Locus |
Thorou- |
Arabian |
Akhal-Teke |
Trakehner |
Orel trotter |
Russian trotter |
Standar- |
|||||||
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
|
AHT4 |
8 |
R |
8 |
– |
9 |
F, Q |
4 |
– |
8 |
L |
5 |
– |
5 |
– |
AHT5 |
6 |
I |
5 |
– |
5 |
– |
6 |
Q |
6 |
– |
5 |
– |
5 |
– |
ASB2 |
12 |
U |
12 |
– |
12 |
G, H |
11 |
– |
5 |
– |
9 |
– |
6 |
– |
ASB17 |
5 |
– |
10 |
– |
10 |
– |
7 |
– |
11 |
I, J |
6 |
– |
10 |
– |
ASB23 |
6 |
– |
8 |
– |
8 |
R |
7 |
V |
6 |
– |
5 |
– |
6 |
– |
CA425 |
6 |
– |
8 |
– |
8 |
– |
3 |
– |
9 |
F |
5 |
– |
6 |
– |
HMS1 |
4 |
– |
5 |
– |
5 |
– |
3 |
– |
5 |
– |
5 |
– |
4 |
– |
HMS2 |
8 |
N |
8 |
– |
8 |
– |
6 |
– |
7 |
– |
7 |
– |
7 |
– |
HMS3 |
7 |
– |
10 |
H, L |
10 |
G, J, S |
6 |
– |
6 |
– |
7 |
– |
6 |
– |
HMS6 |
6 |
– |
7 |
– |
7 |
J |
6 |
– |
5 |
– |
4 |
– |
5 |
I |
HMS7 |
9 |
– |
6 |
– |
7 |
– |
7 |
– |
6 |
– |
4 |
– |
6 |
– |
HTG4 |
8 |
R |
5 |
– |
7 |
– |
5 |
– |
5 |
– |
6 |
– |
5 |
– |
HTG6 |
8 |
N, M |
4 |
– |
6 |
– |
6 |
– |
4 |
– |
5 |
– |
3 |
– |
HTG7 |
4 |
– |
4 |
– |
5 |
P, I |
3 |
– |
3 |
– |
3 |
– |
4 |
– |
HTG10 |
6 |
– |
9 |
– |
8 |
Q |
6 |
– |
8 |
– |
4 |
P |
6 |
P |
LEX3 |
8 |
– |
11 |
G |
9 |
– |
5 |
– |
8 |
– |
5 |
– |
6 |
– |
VHL20 |
8 |
H |
7 |
– |
11 |
J, K, S |
7 |
– |
7 |
– |
7 |
– |
7 |
– |
Total |
104 |
8 |
102 |
3 |
140 |
15 |
100 |
2 |
109 |
4 |
92 |
1 |
97 |
2 |
Note. Nа — number of alleles in a locus, Pа — private alleles (a dash corresponds to the absence of private alleles) |
||||||||||||||
In trotting breeds, the highest polymorphism degree of microsatellite DNA was found in Orlov trotter – it carries four alleles not detected in other breeds. Standardbred trotters (the ancestor - Hambletonian X carrying a significant share of Thoroughbred bloods) were found to have more consolidated allele fund and two private alleles. Russian trotter breed – the result of the complex reproductive crossbreeding of Orlov and American trotters in the middle of last century – have a total set of alleles of microsatellite loci from both source breeds. French trotters are known to carry some of the Standardbred, so DNA of this breed revealed the allele HTG7L. It’s notable that trotters of different breeds were distinct from riding horses by the presence of alleles ASB17F and HMS2P, which possibly indicates the influence of genes of the 10th and 15th chromosomes on genetic determination of fast gait trotting.
2. The genetic spectrum of alleles of microsatellite loci in local horse breeds bred in Russia (n = 1169) |
||||||||||||||
Locus |
Altai |
Buryat |
Vyatka |
Transbaikalian |
Mezen |
Tuvan |
Khakassian |
|||||||
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
Nа |
Pа |
|
AHT4 |
7 |
– |
5 |
– |
8 |
– |
5 |
– |
6 |
– |
7 |
– |
5 |
– |
AHT5 |
7 |
P |
4 |
– |
3 |
– |
6 |
– |
6 |
– |
4 |
– |
6 |
– |
ASB2 |
11 |
– |
5 |
– |
6 |
– |
5 |
– |
6 |
– |
7 |
– |
7 |
– |
ASB17 |
13 |
W, V |
7 |
– |
6 |
– |
7 |
– |
6 |
– |
6 |
– |
7 |
T |
ASB23 |
6 |
– |
6 |
– |
3 |
– |
6 |
– |
6 |
– |
8 |
G |
6 |
– |
CA425 |
8 |
– |
6 |
– |
6 |
– |
6 |
– |
5 |
– |
6 |
– |
6 |
P |
HMS1 |
6 |
– |
4 |
– |
5 |
– |
4 |
– |
4 |
H |
4 |
– |
4 |
– |
HMS2 |
7 |
– |
4 |
– |
7 |
– |
4 |
F |
5 |
– |
5 |
– |
4 |
– |
HMS3 |
7 |
– |
4 |
– |
7 |
– |
4 |
– |
7 |
– |
6 |
– |
6 |
– |
HMS6 |
6 |
– |
4 |
– |
4 |
– |
4 |
Q |
6 |
– |
4 |
– |
4 |
– |
HMS7 |
6 |
– |
4 |
P |
5 |
– |
5 |
– |
5 |
– |
5 |
– |
5 |
– |
HTG4 |
7 |
– |
6 |
– |
8 |
– |
6 |
– |
5 |
– |
5 |
– |
4 |
– |
HTG6 |
5 |
– |
6 |
– |
5 |
– |
6 |
– |
2 |
– |
5 |
– |
4 |
– |
HTG7 |
6 |
– |
4 |
– |
5 |
– |
4 |
– |
4 |
– |
4 |
– |
3 |
– |
HTG10 |
10 |
– |
7 |
– |
5 |
– |
7 |
– |
8 |
– |
7 |
– |
7 |
– |
LEX3 |
11 |
– |
6 |
– |
4 |
– |
6 |
– |
4 |
R |
7 |
– |
7 |
– |
VHL20 |
8 |
– |
7 |
– |
3 |
– |
7 |
– |
6 |
– |
8 |
– |
8 |
– |
Total |
129 |
3 |
91 |
1 |
90 |
0 |
98 |
2 |
92 |
2 |
89 |
1 |
93 |
2 |
Note. See Table 1. |
||||||||||||||
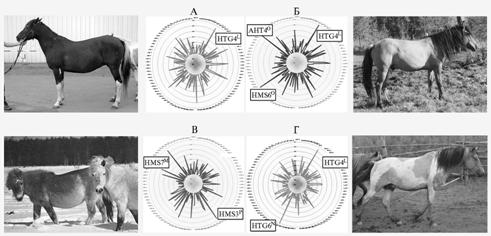
Almost all local breeds showed high polymorphism degree of microsatellite loci even at small number of tested animals. The genetic structure of six breeds includes private alleles not found in plant horses. Two private alleles were detected in Khakassian (ASB17T, CA425P), Mezen (HMS1H, LEX3R) and Transbaikalian (HMS2F, HMS6Q), one - in Buryat (HMS7P) and Tuvan (ASB23G) horses (Table 1). High frequency of the allele HTG4L was observed in Transbaikalian, Tuvan and Khakassian horses (Fig. 1), as well as in Akhal-Teke breed. Vyatka local breed revealed the specific genetic profile and the absence of unique alleles.
Average polymorphism degree of tested breeds varied from 3,2 to 4,5, and plant horses had lower genetic variability than local breeds due to the influence of artificial selection (Table 3). The lowest polymorphism of both microsatellite loci and structural genes was found in Arabian thoroughbred breed.
The degree of heterozygosity (He) for the loci in tested populations averaged from 0,656 (Arabian thoroughbred) to 0,748 (Tuvan). In general, both farm and local breeds showed quite high degree for almost all studied microsatellite loci, which allows using satellite DNA polymorphism for assessment of heterozygosity in individual animals and populations.
High values of the average number of alleles per one locus (NV) were detected in Akhal-Teke (8,2) and Altai (7,5) breeds. In seven populations, the deficit of heterozygous genotypes was reflected by positive inbreeding coefficient (Fis). The excess of heterozygous genotypes was observed in Orlov, Russian and French trotters, Trakehner and Thoroughbred riding horses, and in three local breeds - Altai, Khakassian and Mezen.
|
Fig. 1. Frequency distribution of microsatellite DNA alleles in local horse breeds bred in Russia: A – Tuvan, B – Khakasskaya, C – Buryat, D – Transbaikalian. |
3. Genetic and population characteristics of farm and local horse breeds bred in Russia, analyzed for 17 microsatellite loci |
||||||
Breed |
n |
Ае |
He |
Но |
Fis |
NV |
Altai |
29 |
4,466 |
0,744 |
0,723 |
-0,070 |
7,471 |
Arabian |
319 |
3,213 |
0,656 |
0,610 |
0,061 |
6,824 |
Akhal-Teke |
109 |
3,897 |
0,731 |
0,696 |
0,048 |
8,235 |
Buryat |
11 |
3,609 |
0,701 |
0,748 |
0,052 |
5,294 |
Vyatka |
10 |
3,721 |
0,691 |
0,680 |
0,018 |
5,294 |
Transbaikalian |
11 |
4,013 |
0,729 |
0,765 |
0,019 |
5,820 |
Mezen |
12 |
3,808 |
0,693 |
0,605 |
-0,110 |
5,529 |
Orlov trotter |
99 |
3,501 |
0,685 |
0,710 |
-0,035 |
6,402 |
Russian trotter |
14 |
3,450 |
0,669 |
0,716 |
-0,070 |
5,412 |
Standardbred |
50 |
3,433 |
0,659 |
0,652 |
0,011 |
5,706 |
Trakehner |
27 |
3,861 |
0,706 |
0,670 |
-0,120 |
5,353 |
Tuvan |
11 |
4,204 |
0,748 |
0,776 |
0,013 |
5,882 |
French trotter |
13 |
3,379 |
0,686 |
0,723 |
-0,054 |
5,235 |
Khakassian |
11 |
4,045 |
0,726 |
0,723 |
-0,020 |
5,647 |
Thoroughbred |
443 |
3,519 |
0,689 |
0,697 |
-0,010 |
6,875 |
Note. n — number of animals, Aе — polymorphism degree, Hе — expected homozygocity, Ho — observed homozygocity, Fis — population inbreeding coefficient, NV — average number of alleles per locus. |
||||||
|
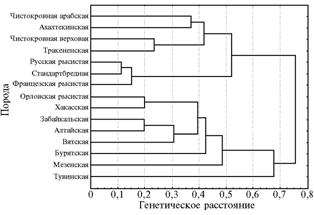
Fig. 2. The dendrogram showing resemblance for 17 microsatellite loci of farm and local horse breeds bred Russia Note: |
The cluster analysis of genetic resemblance of studied breeds resulted in the dendrogram (Fig. 2), that proves the pronounced genetic divergence of two evolutionary branches – farm and local horse breeds. The only exclusion is Orlov trotter shown in the group of local breeds, which is explained rather by its widespread systematic use in improvement of local breeds than by its origin.
The highest coefficient of genetic resemblance of microsatellite DNA loci (0,887) was observed in Russian trotter and a Standardbred horse, which clearly reflects the increasing pur-sang in Russian trotter. These two genetically close species are related to French trotter having a Standardbred admixture. In the subgroup of local breeds, Tuvan horse is a separate branch owing to its historical development in geographic isolation and the resulting low exposure to cross-breeding with other breeds.
Thus, each of the 15 studied horse breeds – farm and local ones – has an individual genetic structure with several private alleles. The highest degree of genetic diversity was observed in horses of ancient cultural breed - Akhal-Teke, the lowest - in the Arabian breed. It has been established the high degree of genetic resemblance of local domestic breeds carrying a wide range of alleles in a single cluster, that indicates their common origin. The detected genetic features of different horse breeds provide the additional information for studying their origin and can be used in programs for preserving gene pools of small populations.
REFERENCES
1. Marklund S., Ellegren H., Eriksson S., Sandberg K. and Andersson L., Parentage Testing and Linkage Analysis in the Horse Using a Set of Highly Polymorphic Microsatellites, Animal Genetics, 1994, vol. 25, pp. 19-23.
2. Binns M.M., Holmes N.G., Holiman A. and Scott A.M., The Identification of Polymorphic Microsatellite loci in the Horse and Their Use in Thoroughbred Parentage Testing, British Vet. J., 1995, vol. 151, pp. 9-16.
3. Bowling A.T. and Ruyinsky A., The Genetics of the Horse, Wallington, UK, 2000.
4. Bjornstad G. and Roed K.H., Breed Demarcation and Potential for Bred Allocation of Horses Assessed by Microsatellites Marker, Animal Genetics, 2001, vol. 32, pp. 59-65.
5. Aberle K.S., Hamann H., Drogemuller C. and Distl O., Genetic Diversity in German Draught Horse Breed Compared with a Group of Primitive, Riding and Wild Horses by Means of Microsatellite DNA Markers, Animal Genetics, 2004, vol. 35, pp. 270-277.
6. Luis C., Juras R., Oom M.M. and Cothran E.G., Genetic Diversity and Relationships of Portuguese and Other Horse Breeds Based on Protein and Microsatellite Loci Variation, Animal Genetics, 2007, vol. 38, pp. 20-27.
7. Dubrovskaya R.M. and Starodumov I.M. Allelefund of Loci for Transferrin, Albumin, Esterase and Blood Groups in Ten Breeds of the Horse Bred in USSR, in Sb. nauch. tr. VNII konevodstva: Rezervy povysheniya effektivnosti konevodstva i konnozavodstva (Compilation of Sci. Works of the All-USSR Research and Development Institute of Horse Breeding: Reserves for Improvement in Horse Breeding and Farming), Divovo, 1987, pp. 55-69.
8. Khrabrova L.A., The Influence of Selection on Genetic Structure of Horse Breeds, in Sb. nauch. tr. VNII konevodstva: Nauchnoe obespechenie konkurentosposobnosti plemennogo, sportivnogo i produktivnogo konevodstva v Rossii i stranakh SNG (Compilation of Sci. Works of the All-Russia Research and Development Institute of Horse Breeding: Science Support for Competitiveness of Horse Breeding in Russia and CIS for Pedigree, Sport and Production Purposes), Divovo, 2007, part 2, pp. 7-12.
9. Khrabrova L.A., Kalinkova L.V. and Zaitseva M.A., Genetic Differentiation of Thoroughbred Horses on Microsatellite Loci, S.-kh. biol.,2008,no. 2, pp. 31-34.
10. Khrabrova L.A. and Zaitsev A.M., Metodicheskie rekomendatsii po vedeniyu geneticheskogo monitoringa mestnykh porod loshadei (Methodological Recommendations on the Performance of Genetic Monitoring of Local Horse Breeds), Divovo, 2005.
All-Russia Research and Development Institute of Horse Breeding, RAAS, Ryazan’ province, Rybnovsky region, Divovo settlement 391105, Russia |
Received October 20, 2010
|