doi: 10.15389/agrobiology.2012.1.14eng
УДК 635.64:631.52:575.22:[573.6.086.83+577.21]
GENOTYPICAL VARIABILITY IN PROGENY OF TRANSGENIC INTERSPECIFIC HYBRID OF TOMATO. II. IDENTIFICATION OF ALIEN DNA INTEGRATION SITE
Yu.V. Chesnokov1, N.I. Bocharnikova2, L.V. Esaulova3, Z.R. Yunusov4
It was done the molecular-genetic analysis of 19 forms of transgenic genotypes of different progenies and two parents forms of interspecific tomato hybrid Lycopersicon esculentum L. (cv. Fakel) × Solanum pennellii Cor. The difference was revealed between lengths of obtained PCR fragments. Alignment of primary structures of PCR amplicons allowed to determine exact site of integration of alien exogenous DNA in genome of transgenic plants. Based of obtained data it was assumed that integration was done by homologous recombination by triplet AGG from 5' end of integrated in genome fragment and by non-homologous recombination in unidentified 3' end nucleotide site.
Keywords: interspecific hybrid of tomato, alien exogenous DNA, integration site.
Foreign DNA transferred into plant cells using one of available methods commonly integrates into the nuclear genome and is being inherited under Mendelian laws (1, 2). Integration sites are randomly distributed in the genome (3, 4), but in most cases exogenous DNA integrates into one locus as a single copy or a cluster of tandemly linked copies (5). During a genetic transformation induced by pathogenic Agrobacterium or using several methods of direct transfer of foreign DNA, the fact of transformation has been proven at levels of morphology, classical genetics, biochemistry and molecular genetics. Most of experiments on genetic transformation through a germinating pollen usually imply only phenotypical and hybridological analysis of obtained forms. All four levels of proof were enabled in such work on Petunia hybrida (transforming agent - DNA of transducing phage with transferase gene of Escherichia coli) (6). In later experiments on germinating pollen, the effect of genetic transformation, as a rule, was demonstrated at any levels (7-10) regardless of an object.
In the authors’ experiments on tomato, genetic transformation of plants has been performed through a natural pollination and fertilization; the fact was supported by morphological, biotechnological, biochemical and molecular genetic data (11-14). It has been demonstrated the successful introduction of transforming agent (foreign exogenous plasmid DNA carrying a selective marker gene nptII determining resistance of transgenic forms to antibiotic kanamycin) into intact tomato plants during pollination-fertilization. Transgenic forms were isolated at early developmental stages in cell culture in vitro (15), and the resulting seed progenies inherited resistance to kanamycin (16).
Until today, it haven’t been performed any research on genetic transformation of plants simultaneous with distant hybridization. In authors’ opinion, the development of such innovative techniques would give much more opportunities to create the improved crops with new economically valuable traits established upon the extended range and frequency of available genotypic diversity. New forms obtained through the joint use of genetic transformation and distant hybridization can be quite useful for understanding the structure and functioning of plant genome, and they also can help to optimize the methods of assessing and selecting valuable transgenic genotypes. The earlier findings (11-14) are important for further development of theoretical ideas about the evolutionary role of genomic variation caused by exogenous and endogenous DNA; these data allow to consider genetic instability as one of key processes providing genotypic variability in natural and artificial populations. At the same time, there is an obvious practical importance of the obtained transgenic and mutant tomato forms (11-14, 16) suitable for direct use in breeding programs as a source material. Along with it, breeding value of resulting forms was evaluated (17).
The purpose of this work was identification of genetic transformation in valuable breeding samples of tomato and their parental forms using molecular genetic analysis and revealing the exact location of exogenous foreign DNA integrated in the genome of selected interspecific hybrids Lycopersicon esculentum Mill. × Solanum pennellii Cor.
Technique. The material for the study were transgenic genotypes of the interspecific tomato hybrid obtained through the cross of a cultivated Lycopersicon esculentum Mill. (cv Fakel) and a wild sample of Solanum pennellii Cor. with exogenous foreign plasmid DNA carrying transgene nptII determining resistance to kanamycin (16).
Total genomic DNA was isolated from fresh or freeze-dried and stored at +4 °C plant material (shredded tissues, leaves, sprouts) using the technique described by K. Lichtenstein and J. Draper (18).
PCR was performed under the following regime: 5 min - denaturation at 94 °C, 1 min – annealing of primers at 50-55 ° C, 1,5 min – polymerization at 72 °C, 1 min - denaturation at 94 °C, 10 min - final polymerization at 72° C; final temperature 4 °C. Each PCR included 35-40 cycles. Taq-polymerase and corresponding buffers were applied according to manufacturer's protocol (“Boehringer”, Germany). Amplification was carried out using the following pairs of primers homologous to conservative regions of legumin-like proteins of Gnetum gnemon, Ephedra gerardiana and vicilin-like protein of Matteucia struthiopteris (19, 20):
G. gnemon— |
GCGGCAGGGCCAATTCCTTCTCATCCCGCAGAACTTCGC, |
E. gerardiana— |
CCGCAGTTCTTCGCTGCTGTGTAAAAGAGGCCTCCGAAGAAGG, |
M. struthiopteris— |
GCCTTACAACCTATTCAAAGAGAAGCCGGATTTTGGC, |
Resulting PCR products were sequenced using ABI PRISMTM system (“Perkin Elmer”, USA) under the protocol for a sequencing set ABI PRISMTM Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA Polymerase FS (P/N 402078) from the same manufacturer. Horizontal electrophoresis of DNA fragments was performed in 1% agarose gel in TBE or TAE buffer and electric field of 2,4 V/cm. After electrophoresis, the gel was stained with ethidium bromide at a concentration of 0,5 ug/ml. Results of PCR amplification and gel electrophoresis were visualized using a system BioDoc It (“UVP”, UK).
For electroelution, DNA from 1% agarose gel after electrophoresis in TAE buffer at 3 V/cm and staining with ethidium bromide, a region of gel containing desired DNA fragment was excised and transferred to a dialysis tube with TAE buffer. The tube was sealed on both sides, placed in electrophoresis chamber and subject to a voltage 4,5 V/cm. After the release of DNA from the gel, a direction of current was changed several times for periods of 5-10 c. Then the gel was removed and the tube contents were concentrated up to 300-400 ul and transferred to a centrifuge tube of 1,5 ml volume (“Eppendorf”, Germany). DNA solution was treated once with an equal volume of phenol-chloroform mixture (1:1). The upper water phase was isolated and added with NaCl to a content 0,1 M; DNA was precipitated using two volumes of chilled ethanol while leaving the tube for 2-3 h at –20 °C. The precipitated DNA was collected by centrifugation for 10-15 min at +4 °C. The precipitate was washed with 75% ethanol, slightly dried and dissolved in 20 ml bidistilled water. Resulting DNA solution was stored at - 20 C°.
In order to establish a possible nature of analyzed DNA fragments, the data on homologous nucleotide sequences were observed in known databases using interactive program BLAST Search available the Internet (http://www.ncbi.nim.nih.gov/cgi- bin / BLAST / nph-newblast? Jform = 1).
Results. PCR analysis was performed on total genomic DNA isolated from transgenic interspecific hybrids of tomato which had been obtained previously (17). Knowing that transgenic DNA can affect the recipient’s genome already in early or late embryogenesis, as PCR primers there were used three pairs of oligonucleotide primers homologous to conservative amino acid fragments of seed storage proteins of G. gnemon, E. gerardiana and a vicilin-like protein from spores of M. struthiopteris (19, 20). In contrast to all other known species (probably, except only Physarum polycephalum), these organisms were described as ones having the evolutionarily most ancient, “inferior” proteins related to a superfamily of structurally homologous but functionally distinct products (21).
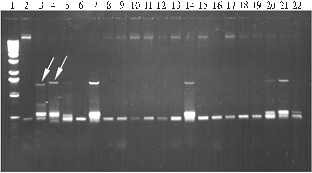
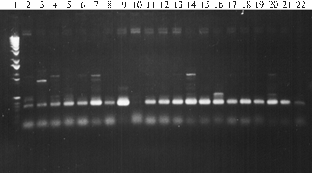
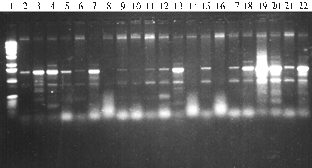
The analysis revealed a dissimilarity of PCR spectra of amplified fragments in both parental forms and their transgenic hybrid progenies (Fig. 1). Thus, in the case of primers G. gnemon, there were detected several fragments (from 400 to 1500 bp), some of which were peculiar to all tested genotypes including parental forms, while others were unique and specific only to certain transgenic forms. Similar results were obtained in PCR with primers E. gerardiana and M. struthiopteris. However, the number of detected amplified fragments for primers G. gnemon and M. struthiopteris was lower than for primers E. gerardiana.
The fragments of about 1500 bp, e.g. in genotype 2-83 (2) (Fig. 1) in the variant with primers G. gnemon, were the most unique relative to other fragments amplified using both these primers and primers M. struthiopteris and E. gerardiana; such unique fragments were chosen for subsequent molecular-genetic analysis. Sequencing of these fragments was performed using the same primers G. gnemon as in PCR. Along with it, the fragment typical to S. pennellii (approximately 1300 bp) was sequenced simultaneously. Resulting primary nucleotide sequences of 1536 and 1316 bp lengths were subjected to molecular comparison (Fig. 2). Similarity amounted to 85,7% and 220 non-homologous nucleotides were found to be arranged as a single fragment.
A |
|
B |
|
C |
|
Fig. 1. PCR-spectra of total genomic DNA from certain transgenic hybrid genotypes of tomato and their parental forms with primers Gnetum gnemon (A), Ephedra gerardiana(B) and Matteuciastruthiopteris (C): 1 — molecular weight marker (100 bp-1000 bp); 2 —Lycopersicon esculentum; 3 — Solanum pennellii; 4, 7, 14, 20 and 21 — respectively, F2, F3, F5, F7 and F8 of genotype 2-83(2); 5, 8, 10, 12, 15, 18 and 22 —respectively, F2-F8 of genotype 28-26; 6, 9, 11, 13, 17 and 19 —respectively, F2-F7 of genotype 2-83(4). |
In order to identify a possible nature of this fragment, the data on homologous sequences were observed in available databases. It was found 253 sequences somewhat homologous to 1536 bp fragment and 28 sequences similar to 1316 bp fragment of S. pennellii. Among 253 sequences homologous to the amplicon of 1535 bp, 222 ones were described as segments of plasmid vectors (including those intended for transformation of plants) containing the sequence of nptII gene from transposon Tn5 E. coli (registration identity number U32991), and the rest of those were detected in both animals (drosophila, mouse, human) and plants (umbrella liverwort, arabidopsis, tobacco, tomato, etc.). The sequence 1316 bp happened to be homologous only to DNA of animal and plant nature without similarity to any vectors and transposons. Besides, among the 28 abovementioned sequences homologous to it, 19 were also homologous to the fragment 1536 bp. One of these 19 sequences is of the greatest interest – the sequence of clone AJ3 (AF072531) of paracentromeric region of S. pennellii, which is the most homologous to both amplicons of 1536 and 1316 bp.
|
|
Fig. 2. Results of comparative analysis of primary nucleotide sequences of PCR-amplicons 1536 bp (GNETPCR) and 1316 bp (SPENNEL) indicated by arrows on Fig. 1 (А). A triplet of assumed homologous recombination is underlined, nptII fragment of gene - in italics. Numbers indicate location of nucleotides. |
Several nucleotide sequences homologous to both fragments of 1536 bp and 1316 bp are of interest: a sequence for native (AF000999-AF001003, U23053) and modified (U70480, U70481) polygalacturonase of L. esculentum, a fragment of mRNA sequence for legumin-like protein of G. gnemon (Z50779), mRNA of RPB2 gene of Marchantia polymorpha (AF020844), FUS6 gene of Arabidopsis thaliana (L26498), as well as SUA5, PMR1, and tRNA-lys1 genes located in the left arm of the 7th chromosome of Saccharamyces cerevisiae (X85757).
Certain sequences were not homologous to both 1516 bp and 1316 bp amplicons, but to one of these fragments. For example, a nucleotide sequence of Nicotiana sp. (X55365) showing the properties of promoter in transgenic plants and homologous to the fragment 1316 bp. An interesting fact: 222 of 253 sequences containing nptII gene have a similar beginning triplet AGG, which is the ending triplet in the paracentromeric sequence of S. pennellii (AF072531).
Thus, the data of BLAST Search Analysis and sequencing of PCR amplicons allow to conclude that the difference of 220 bp length between the studied fragments 1536 and 1316 bp corresponds to exogenously integrated foreign DNA segment containing a structural part of the nucleotide sequence of nptII gene from transposon Tn5. This integration most likely occurred through a homologous recombination by AGG triplet from 5' end of the integrated fragment and non-homologous recombination by an unidentified site on its 3'-end. This assumption is reasonably suggested by the presented data of molecular-genetic and comparative analysis of primary sequences of amplicons. However, the exact mechanism of integration of foreign exogenous DNA into the recipient’s genome still requires further research.
REFERENCES
1. De Block M., Herrera-Estrella L., Van Momtagu M. et al., Expression of Foreign Genes in Regenerated Plants and in Their Progeny, EMBO J., 1984, no. 3, pp. 1681-1689.
2. Feldmann K.A. and Marks M.D., Agrobacterium-Mediated Transformation of Germinating Seeds of Arabidopsis thaliana: a Non-Tissue Culture Approach, Mol. Gen. Genet., 1987, vol. 208, pp. 1-9.
3. Chyi Y.-S., Jorgensen R.A., Goldstein D. et al., Location and Stability of Agrobacterium-Mediated T-DNA Insertion in the Lycopersicon Genome, Mol. Gen. Genet., 1986, vol. 204, pp. 64-69.
4. Wallroth M., Gerats A.G.M., Rogers S.G. et al., Chromosomal Location of Foreign Genes in Petunia hybrida. Mol. Gen. Genet., 1986, vol. 202, pp. 6-15.
5. Moraus A., Saul M.W., Essad S. and Potrycus I., Localization by in Situ Hybridization of a Low Copy Chimaeric Resistance Gene Introduced into Plants by Direct Gene Transfer, Mol. Gen. Genet., 1987, vol. 207, pp. 204-209.
6. Hess D., Pollen-Based Techniques in Genetic Manipulation, Int. Rev. Cytol., 1987, vol. 107, pp. 367-395.
7. Hess D., Dressler K. and Nimmrichter R., Transformation Experiments by Pipetting Agrobacterium into the Spikelets of Wheat (Triticumaestivum L.), Plant Sci., 1990, vol. 72, pp. 233-244.
8. Chesnokov Yu.V. and Korol’ A.B., Transfer of Alien Genes into Intact Plants of Maize by Means of the Pollination/Fertilization Process, Genetika, 1993, vol. 29, no. 8, pp. 1345-1355.
9. Chesnokov Yu.V. and Korol’ A.B., Inheritance of the Trait of Kanamycin Resistance in the Genetic Transformation of Maize, Genetika, 1993, vol. 29, no. 8, pp. 1492-1499.
10. Smith C.R., Saunders J.A., Van Wert S. et al., Expression of GUS and CAT Activities Using Electrotransformed Pollen, Plant Sci., 1995, vol. 104, pp. 49-58.
11. Bocharnikova N.I., Senyuk V.I., Chesnokov Yu.V. and Kozlova V.M., Evaluation of the Electrophoretic Protein Patterns in Forms of Tomato Mutant for the Trait of Inflorescence Type, Izv. AN RM, Ser. biol. i khim. nauk, 1993, no. 5, pp. 72-73.
12. Bocharnikova N.I., Chesnokov Yu.V. and Kozlova V.M., Effects of Exogenous DNA Action in Tomato. II. Genetic Variability, Genetika, vol. 31, no. 5, pp. 692-696.
13. Vikonskaya N.A., Bocharnikova N.I. and Chesnokov Yu.V., Phenotypic Evaluation of Characters in F2 Segregating Populations of an Interspecific Hybrid of Tomato, Izv. AN RM, Ser. biol. i khim. nauk, 1994, no. 5, pp. 72-73.
14. Chesnokov Yu.V., Sedov G.I. and Vikonskaya N.A., Effects of Exogenous DNA Action in Tomato. I. Genetic Transformation, Genetika, 1995, vol. 31, no. 5, pp. 648-691.
15. Chesnokov Yu.V. and Sedov G.I., In Vitro Selection of Tomato Transformants at the Immature Embryo Stage, Euphytica, 1995, vol. 81, no. 1, pp. 79-83.
16. Chesnokov Yu.V., Genetic Transformations Induced by the Transfer of Exogenous DNA in Higher Plants by Means of Germinating Pollen, Doctoral Sci. Dissertation, St. Petersburg, 2000.
17. Chesnokov Yu.V., Bocharnikova N.I. and Esaulova L.V., Genotypic Variability in Progeny of Transgenic Interspecific Hybrid of Tomato. Communication I. Isolation of Selective Material, S.-kh. biol., 2011, no. 1, pp. 60-65.
18. Lichtenstein C.P. and Draper J., Genetic Engineering of Plants. DNA Cloning: a Practical Approach, Moscow, 1988, pp. 315-380.
19. Shutov A.D., Braun H., Chesnokov Yu.V., Horstmann Ch., Kakhovskaya I.A. and Baumlein H., Sequence Peculiarity of Gnetalean Legumin-Like Seed Storage Proteins, J. Mol. Evol., 1998, vol. 47, no. 4, pp. 486-492.
20. Shutov A.D., Braun H., Chesnokov Yu.V. and Baumlein H., A Gene Encoding a Vicilin-Like Protein is Specifically Expressed in Fern Spores. Evolutionary Pathway of Seed Storage Globulins, Eur. J. Biochem., 1998, vol. 252, no. 1, pp. 79-89.
21. Chesnokov Yu.V., Molecular Evolution of Plant Seed Globulins, Dokl. RASKhN, 2009, no. 4, pp. 8-12.
1N.I. Vavilov All-Russia Research and Development Institute of Plant Growing, RAAS, St. Petersburg 190000, Russia, |
Received 10.05.2011 |